Final report for GNE19-196
Project Information
Oocyte quality remains an undefined obscure topic in reproductive research. The goal of this research was to take a new approach to tackle this decades-old problem. As technology has increased more and more aspects of everyday life have started to utilize technology and take a turn more towards science. The goal was to incorporate molecular physiology with whole animal applications by investigating long noncoding RNAs (lncRNAs) and their potential roles in bovine reproduction specifically, oocyte quality. In the first year and a half of funding, I successfully verified the presence and differential expression of six highly abundant lncRNAs in single bovine oocytes aspirated from small and large follicles at varying developmental stages. More specifically, I documented the significance of the maturity stage in all six lncRNAs and graphically showed their accumulation and degradation during maturation. When examining each for the interaction of follicle size and maturity stage, there was a significant interaction in the expression of OOSNCR5 (P < 0.05) and a tendency for significant interaction in the expression of OOSNCR2. The literature indicates that follicle size can be an indicator of oocyte quality and that RNAs accumulated or degraded during maturation are crucial to achieving developmental competence. Therefore, lncRNAs OOSNCR2 and OOSNCR5 show promise to be linked with oocyte quality.
My second year and a half of funding were met with challenges. As the pandemic continued to plague our world, researchers like everyone else have learned to roll with the punches and push forward. Due to travel restrictions and various shutdowns, our lab had to shut down the in vitro embryo production portion of our lab. With the shutdown, we were unable to collect ovaries from the abattoir completely halting all projects. After a year and six months, we were finally approved to travel again. Sample collections began in September 2021.
In the last three months, we traveled bi-weekly to the commercial abattoir in hopes of producing data again. I successfully produced and quantified a tissue expression profile, verifying the oocyte-specificity of three lncRNAs and possibly an additional two. Various tissue samples (n = 26) were collected from freshly slaughtered beef cattle. Real-time qPCR analysis was performed and lncRNA expression was examined using a one-way ANOVA followed by Tukey's HSD post hoc test. In addition, follicular cells; cumulus, granulosa, and theca cells were collected from individual follicles. Follicular cells were subjected to qPCR analyzed using a one-sided t-test. When investigating lncRNA expression in various tissues, three lncRNAs were expressed solely in oocytes [fetal ovaries] with one lncRNA being detected in the adult ovary and one showing variable expression. Minimal expression was detected in the granulosa and theca cells. LncRNA expression in the cumulus cells (not included in the presented data) exhibits patterns similar to those seen in the maturity stage of single oocytes. Based on these data, we conclude lncRNAs OOSNCR1, OOSNCR3, and OOSNCR5 are oocyte-specific. Future investigations will focus on characterizing the mechanisms for the three oocyte-specific lncRNAs and their roles in oocyte maturation and early embryonic development.
Due to time constraints, objective 1 was unable to be completed. In situ hybridization is scheduled to start in the very near future. With regards to objective 3, it was unable to be completed, again due to time constraints and lack of sample collections for over a year. All supplies have been purchased and proper training has gotten underway to perform microinjection to elucidate the functional mechanisms utilized by specific lncRNAs. In the last month, preliminary data has been collected for three additional studies.
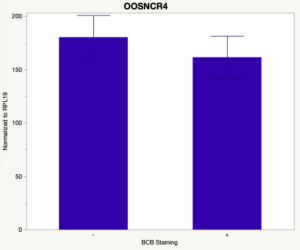
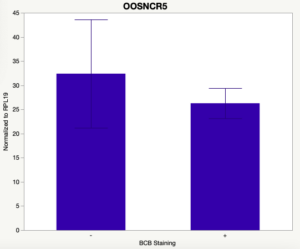
The first study utilized Brilliant Cresyl Blue (BCB) staining for the selection of immature oocytes before in vitro embryo production. Previously, oocytes have been selected as "good or bad" based on morphological assessment including the compactness and number of cumulus cells surrounding the oocyte, follicle size, cytoplasmic morphology, and oocyte size. All of which have proven to be highly subjective and difficult to replicate across laboratories. The need for a standardized measure of oocyte quality is more than obvious. However, a standardized measurement remains elusive. It is widely believed that glucose-6-phosphate dehydrogenase (G6PDH) is actively produced in growing oocytes, but its activity is decreased in oocytes that have finished their growth and achieved developmental competence. The enzyme G6PDH can degrade BCB thus fully grown [competent] oocytes show a blue cytoplasm (BCB+) after BCB staining while growing [immature] oocytes have high levels of G6PDH and exhibit a colorless cytoplasm (BCB-) after staining. BCB has previously been published in a wide variety of species as a marker of oocyte maturity. It has been published that BCB+ oocytes display a higher blastocyst developmental rate than BCB- and control oocytes in pig (Roca et al., 1998), goat (Rodriguez-Gonzalez et al., 2002)), sheep (Catala et al., 2011), mouse (Wu et al., 2007), dog (Rodrigues et al. 2009), and bovine (Bhojwani et al., 2007). Preliminary data suggests lncRNA expression displays an opposite pattern than expected with lower lncRNA expression in the BCB+ oocytes. Possibly suggests the selected lncRNA are necessary for oocyte growth. The data associated with BCB staining will be used as a puzzle piece that hopefully will help elucidate the functions of these unknown genes.
The second preliminary study conducted was to determine if the specific lncRNAs are maternal transcripts. During early embryogenesis, maternal mRNAs that accumulated in the oocyte during oogenesis, play important roles in embryonic genome activation (Hamatani et al., 2004). Some maternal transcripts are oocyte-specific and known as maternal effect genes which are required for the early cleavage events post-fertilization. Alpha amanitin is a selective inhibitor of RNA polymerase II. By culturing embryos in the presence of alpha amanitin maternal transcripts can be differentiated from embryonic transcripts. Preliminary data suggests a complicated relationship and a possible negative regulation by a maternal transcript.
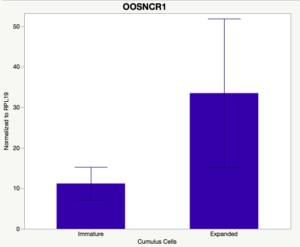
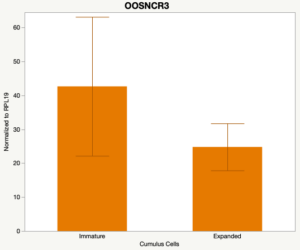
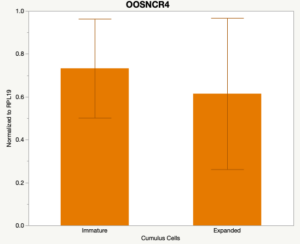
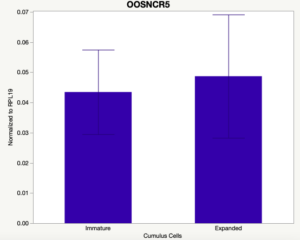
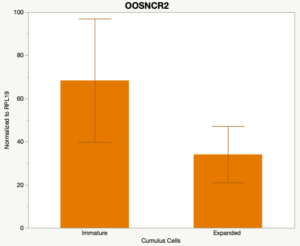
The third and final study on the horizon is to further investigate the bidirectional communication between the oocyte and its surrounding nurse [cumulus] cells. With the recent focus on transzonal projections (TZPs), there is compelling data to suggest that these cellular projections may be involved in the transporting and delivery of large molecules to the oocyte. It is well accepted that endogenous transcription of the oocyte is silent during oocyte maturation. Accumulation of transcripts in the oocyte during this time would support the potential transfer of material from the cumulus cells to the oocyte. Preliminary data collected under specific aim 2 supports the accumulation of predicted lncRNAs occurred during oocyte maturation. Those that were accumulated provide valid reasoning for selection to investigate their roles in the bidirectional communication between the cumulus cells and the oocyte. Data [unpublished] collected from immature and expanded cumulus cells display identical lncRNA expression accumulation and degradation patterns to those seen in single oocytes at corresponding maturity stages. Although preliminary, these data suggest a possible relationship between the two cell types that require further investigation.
This experiment may seem like a stretch for sustainable agriculture, especially discussing gene expression data. However, studies of this nature are the first steps necessary in determining potential bio-markers that can one day be utilized to predetermine which oocytes will produce viable embryos. By screening oocytes and embryos prior to fertilization and transfer farmers will be able to select the highest quality gametes. Once the basic research has been conducted, optimization techniques can be performed to translate what we've learned in the lab into a practical farm application. Although we will never fully understand the mechanism of how life begins, well-characterized biomarkers give both farmers and veterinarians the best possible scenario to invest their money in.
1. To characterize the temporal and spatial expression of oocyte-specific lncRNAs during early embryogenesis and determine if they are involved in fertilization.
2. To investigate oocyte quality by comparing lncRNA expression profiles in “good and bad” oocytes based on previously published methodologies; follicle size and cytoplasmic morphology.
3. To elucidate the regulatory role of specific lncRNAs in meiotic maturation, fertilization, cleavage, and developmental events characteristic to the maternal-to-embryonic transition.
4. To characterize the bidirectional communication of specific lncRNAs during oocyte maturation via the transzonal projections.
The purpose of my project was to make novel discoveries investigating the potential roles of long noncoding RNAs in bovine reproduction. Up to this point in time, oocyte quality remains difficult to define. My goal was to approach it from a different perspective. Molecular genetics were used to screen oocytes of different qualities to try to find similarities between gene expression and oocyte quality. Although this research may seem too far away from sustainable agriculture, I believe that thinking in itself is the problem. I believe that the future of sustainable agriculture lies in the utilization of technology. There comes a point when farmers and scientist must work together to tackle the issues seen among their herds. Scientist have the scientific knowledge and the means to make great new discoveries while the farmers have the agricultural knowledge and the animals needed to mesh the two fields. I believe my research is important for the future of sustainable agricultural because it takes the new generation of thinking to combine science with hundreds of years of agricultural knowledge. I do not have the means or knowledge to run a farm, however I know how to perform in vitro embryo production which when used properly, has the means to produce genetically superior animals that can yield hundreds of thousands of dollar worth of profit for a farmer. By combining a scientists knowledge with an open minded farmer who's willing to try something new. I believe that is the future of sustainable agriculture.
Research
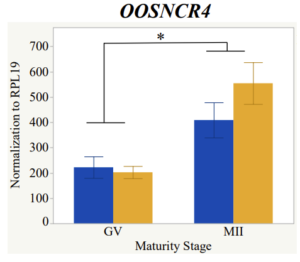
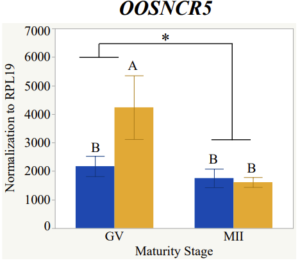
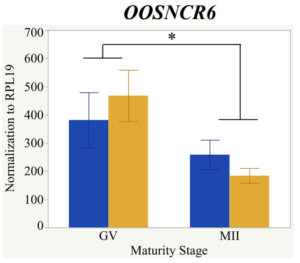
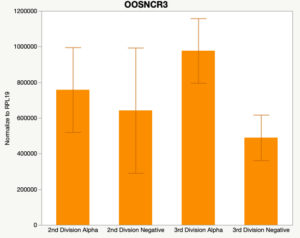
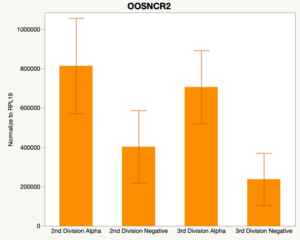
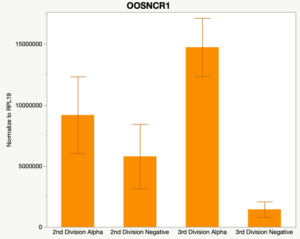
With the past year and a half of funding I have completed objective 2.) To investigate oocyte quality by comparing lncRNA expression profiles in "good and bad" oocytes based on previously published methodologies, follicle size and cytoplasmic morphology. Although I started this objective when I received funding and it was reported in last year's project report, due to the pandemic this year my research was placed on hold for a majority of the year. The break in my lab work allowed me to go back over my data and correct some errors. The originally submitted data was added to and re-analyzed. The methodology remained the same and is as followed. Our laboratory identified 1535 lncRNAs in bovine oocytes using RNA-sequencing (Table 1 below). The objective of this study was to verify our RNA-seq data and if present, characterize the expression of six highly abundant lncRNAs (OOSNCR1, OOSNCR2, OOSNCR3, OOSNCR4, OOSNCR5, and OOSNCR6) in single oocytes aspirated from varying size follicles at different developmental stages. Single oocytes were aspirated from small (<4 mm) and presumably oestrogen active (6–18 mm) follicles and were denuded at the germinal vesicle (GV) or MII stage. Single oocytes were aspirated from small (<4 mm) (SF) and presumably estrogen active (6–18 mm) (EA) follicles and were denuded at the germinal vesicle (GV) or MII stage. MII stage was determined by cumulus expansion and the extrusion of the first polar body. Real-time quantitative PCR analysis was performed, using RPL-19 as an endogenous control for data normalization analyzed using the standard curve method. Average expression of each lncRNA from SF was used as a calibrator in determination of fold change. Effect of follicle size (SF, EA) and maturity stage (GV and MII) and their interaction on the lncRNA expression were examined using a two-way factorial ANOVA followed by Tukey’s HSD post hoc test.
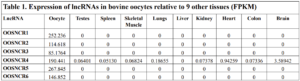
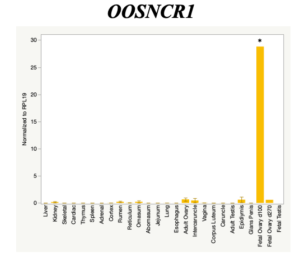
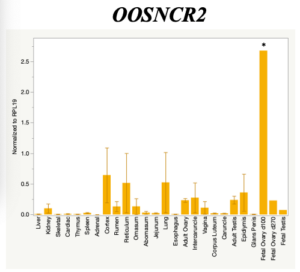
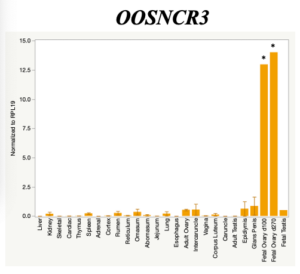
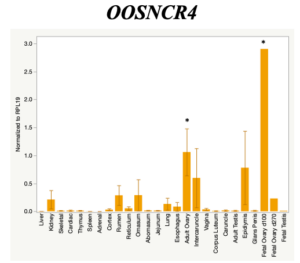
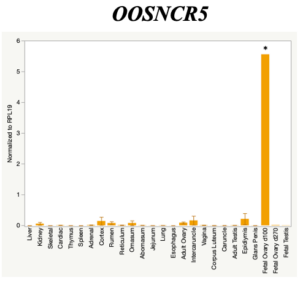
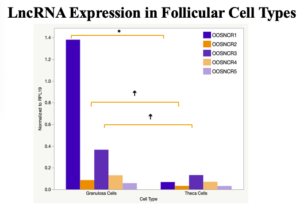
The objective of the second study was to generate tissue and cell-type expression profiles harvested from beef cattle in order to verify the oocyte-specificity of these novel lncRNAs as validation for selection for experimentation. Tissue samples were collected from the liver, kidney, skeletal muscle, cardiac muscle, thymus, spleen, adrenal gland, brain, rumen, reticulum, omasum, abomasum, jejunum, lung, uterus (caruncle and intercaruncle), vagina, adult ovary, corpus luteum, epididymis, glans penis, adult testis, fetal ovary (d100 and d270), and fetal testis. All samples were snap-froze in liquid nitrogen at the abattoir and stored at -800C until isolated. RNA isolation was performed using the Trizol method and cDNA was synthesized using the high capacity cDNA reverse transcriptase kit. RT-qPCR was performed to quantify lncRNA expression. RPL-19 was used as a housekeeping gene and relative gene expression was calculated using the standard curve method. Gene expression data revealed oocyte-specificity in five (OOSNCR1, OOSNCR2, OOSNCR3, OOSNCR4, OOSNCR5) of the eleven lncRNAs investigated. Real-time qPCR analysis was performed and lncRNA expression was examined using a one-way ANOVA followed by Tukey's HSD post hoc test. In addition, follicular cells; cumulus, granulosa, and theca cells were collected from individual follicles. Follicular cells were subjected to qPCR analyzed using a one-sided t-test.
Follicle Size Study
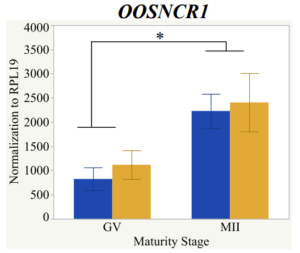
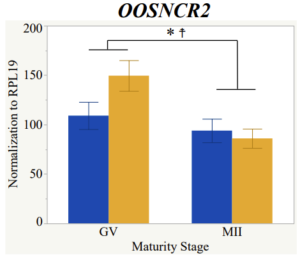
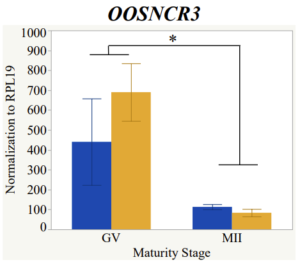
Maturity stage was significant (P < 0.05) in all 6 lncRNAs. OOSCNR2, OOSNCR3, OOSCNR5, and OOSNCR6 were all higher in the GV oocytes and were degraded with maturation. OOSNCR1 and OOSNCR4 displayed opposite expression patterns in that they were accumulated during maturation. When examining each for the interaction of follicle size and maturity stage, there was a significant interaction in the expression of OOSNCR5 (P < 0.05). Within it, SF oocytes at the GV stage had the highest relative expression. A significant difference was detected between SF MII and SF GV oocytes, indicating a higher expression in the earlier developmental stage (P < 0.05). SF oocytes at MII had a 0.63-fold decrease in their expression relative to the SF oocytes at GV. In addition, there was a tendency for significant interaction between stage and size in the expression of OOSNCR2 (P < 0.1). OOSNCR2 exhibited a similar expression pattern as OOSNCR5. Although not significant, SF oocytes at the GV stage had the highest relative expression, indicating a higher expression in the earlier developmental stage. Here the SF oocytes at MII had a 0.43 -fold decrease in their expression relative to the SF oocytes at GV.
Tissue Expression Profile
When investigating lncRNA expression in various tissues, three lncRNAs were expressed solely in oocytes [fetal ovaries] with one lncRNA being detected in the adult ovary and one showing variable expression. Minimal expression was detected in the granulosa and theca cells. LncRNA expression in the cumulus cells (not included in the presented data) exhibits patterns similar to those seen in the maturity stage of single oocytes however, more samples are required.
Unpublished preliminary data
In the last month, preliminary data has been collected for three additional studies.
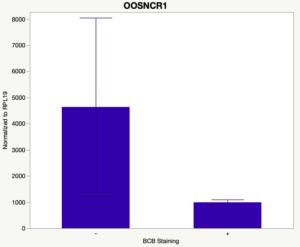
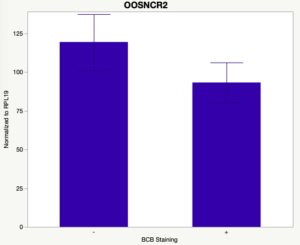
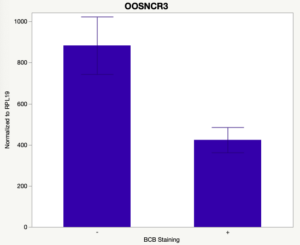
The first study utilized Brilliant Cresyl Blue (BCB) staining for the selection of immature oocytes before in vitro embryo production. Previously, oocytes have been selected as "good or bad" based on morphological assessment including the compactness and number of cumulus cells surrounding the oocyte, follicle size, cytoplasmic morphology, and oocyte size. All of which have proven to be highly subjective and difficult to replicate across laboratories. The need for a standardized measure of oocyte quality is more than obvious. However, a standardized measurement remains elusive. It is widely believed that glucose-6-phosphate dehydrogenase (G6PDH) is actively produced in growing oocytes, but its activity is decreased in oocytes that have finished their growth and achieved developmental competence. The enzyme G6PDH can degrade BCB thus fully grown [competent] oocytes show a blue cytoplasm (BCB+) after BCB staining while growing [immature] oocytes have high levels of G6PDH and exhibit a colorless cytoplasm (BCB-) after staining. BCB has previously been published in a wide variety of species as a marker of oocyte maturity. It has been published that BCB+ oocytes display a higher blastocyst developmental rate than BCB- and control oocytes in pig (Roca et al., 1998), goat (Rodriguez-Gonzalez et al., 2002)), sheep (Catala et al., 2011), mouse (Wu et al., 2007), dog (Rodrigues et al. 2009), and bovine (Bhojwani et al., 2007). Preliminary data suggests lncRNA expression displays an opposite pattern than expected with lower lncRNA expression in the BCB+ oocytes. Possibly suggests the selected lncRNA are necessary for oocyte growth. The data associated with BCB staining will be used as a puzzle piece that hopefully will help elucidate the functions of these unknown genes.
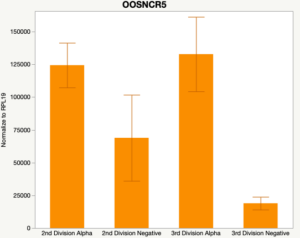
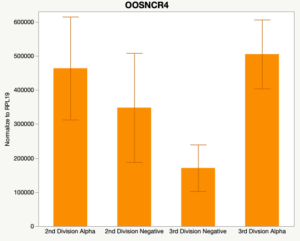
The second preliminary study conducted was to determine if the specific lncRNAs are maternal transcripts. During early embryogenesis, maternal mRNAs that accumulated in the oocyte during oogenesis, play important roles in embryonic genome activation (Hamatani et al., 2004). Some maternal transcripts are oocyte-specific and known as maternal effect genes which are required for the early cleavage events post-fertilization. Alpha amanitin is a selective inhibitor of RNA polymerase II. By culturing embryos in the presence of alpha amanitin maternal transcripts can be differentiated from embryonic transcripts. Preliminary data suggests a complicated relationship and a possible negative regulation by a maternal transcript.
The third and final study on the horizon is to further investigate the bidirectional communication between the oocyte and its surrounding nurse [cumulus] cells. With the recent focus on transzonal projections (TZPs), there is compelling data to suggest that these cellular projections may be involved in the transporting and delivery of large molecules to the oocyte. It is well accepted that endogenous transcription of the oocyte is silent during oocyte maturation. Accumulation of transcripts in the oocyte during this time would support the potential transfer of material from the cumulus cells to the oocyte. Preliminary data collected under specific aim 2 supports the accumulation of predicted lncRNAs occurred during oocyte maturation. Those that were accumulated provide valid reasoning for selection to investigate their roles in the bidirectional communication between the cumulus cells and the oocyte. Data [unpublished] collected from immature and expanded cumulus cells display identical lncRNA expression accumulation and degradation patterns to those seen in single oocytes at corresponding maturity stages. Although preliminary, these data suggest a possible relationship between the two cell types that require further investigation.
BCB Study
Alpha Amanitin Study
Cumulus Cell Study
Due to the nature of this study, the data is very preliminary. Based on the data from the tissue and follicular cell expression profile, we conclude lncRNAs OOSNCR1, OOSNCR3, and OOSNCR5 are oocyte-specific. When paired with the follicle size data, lncRNAs OOSNCR2 and OOSNCR5 show promise to be linked with oocyte quality, future investigations will focus on characterizing the mechanisms for the two oocyte-specific lncRNAs and their roles in oocyte maturation and early embryonic development. By characterizing lncRNAs associated with oocyte quality, one day we hope to be able to discover bio-markers of quality that allows farmers to test for "good" oocytes prior to spending the money necessary to perform in vitro embryo production. Farmers not only would spend less money by only submitting the "good" oocytes to undergo the expensive IVP procedure, but also they would be more willing to utilize the technology knowing it has a good outcome and removing the potential risk of failure.
Education & Outreach Activities and Participation Summary
Participation Summary:
IEST - Follicle Size - Jan 2021
The data provided here was originally submitted as an abstract for the 19th International Congress on Animal Reproduction (ICAR2020) in Bologna, Italy. However, due to the pandemic, the conference was canceled and pushed back to 2022. With the canceling of the conference, all abstracts were discarded and I was able to reanalyze my data and submit it to a different conference. This data will now be presented as a poster at the virtual 47th Annual Conference of the IETS (International Embryo Transfer Society) on January 19th, 2021. This next semester I plan on continuing my research into oocyte quality looking at lncRNA expression as well as tackling portions of objective 1, characterizing the temporal and spatial expression of key lncRNAs. The lncRNAs found in objective 2 will be used for further examination during objective 1. Assuming all goes according to plan, I will be submitting that data as an abstract for the 54th Annual Meeting organized by the Society for the Study of Reproduction (SSR). If accepted, I will be traveling to St. Louis, MO to present my findings.
SSR - Tissue Expression - Dec 2021_SARE2
The data provided here will be presented as a poster at SSR's 54th Annual Meeting (Society for the Study of Reproduction) on December 17th, 2021 in St. Louis, MO. Future studies will include in situ hybridization to localize lncRNA expression, knock out of specific lncRNAs to elucidate functional mechanisms, and lentivirus transfection of cumulus cells to knock out gene expression to study bidirectional communication between the oocyte and surrounding cumulus cells.
Project Outcomes
So far, we are in our preliminary steps and it is too soon for our research to have an affect on agricultural sustainability.
Due to the nature of this project the data is still very preliminary, however, it is very exciting to see oocyte-specificity, differential gene expression in single oocytes, and patterns emerging in cumulus cells, BCB stained oocytes, and alpha amanitin treated embryos. We hope to continue on our path of looking into these long noncoding RNAs as we continue to fit the puzzle pieces together. Our ultimate goal is to use our findings to discover potential whole animal applications. I hope to continue my work in the lab so that eventually I can start to translate my data into something farmers can actually utilize and understand. At the end of the day I hope that my research contributes to bovine reproduction and ultimately, helps to get healthy calves on the ground.
Information Products
- Differential gene expression of bovine long noncoding RNAs in single oocytes aspirated from small and large follicles (Conference/Presentation Material)
- Bovine Tissue Expression Profiles for Five Oocyte-Specific Long non-coding RNAs (Conference/Presentation Material)