Final report for FW20-369
Project Information
Small-scale livestock farmers are looking for innovative ways to manage their lands for environmental sustainability and economic profitability, while ensuring animal well-being. Multi-species rotational grazing (MSRG) has been suggested as a promising approach, since it allows high grazing intensity and stocking density without negatively impacting pasture quality or animal health. However, the potential effects of MSRG on soil microbial communities are poorly understood. To address this knowledge gap, we collected soil microbial samples before, during, and after one season of MSRG by cattle, sheep, and swine. Nine paddocks experiencing MSRG, as well as three paddocks experiencing monospecies grazing (one paddock per species) and one with no grazing, were sampled. Soil samples were sent to the University of Oregon Genomics & Cell Characterization Core Facility for shotgun metagenomic analysis to identify taxonomic, phylogenetic, and functional characteristics of the microbial communities from each paddock.
Results demonstrated broad microbial diversity in all paddock soil microbiomes. Samples collected early in the season tended to have greater archaeal and bacterial alpha diversity than samples collected later for all grazing treatments, while no effect was observed for fungi or viruses. Beta diversity, however, was strongly influenced by both grazing treatment and month for all microbial kingdoms, suggesting a pronounced effect of different livestock on microbial composition. Cattle-only and swine-only paddocks were more dissimilar from multi-species paddocks than those grazed by sheep. We identified a large number of differentially abundant taxa driving community dissimilarities, including Methanosarcinaspp., Candidatus Nitrocosmicus oleophilus, Streptomyces spp., Pyricularia spp., Fusarium spp., and Tunggulvirus Pseudomonas virus phi-2. In addition, a wide variety of antibiotic resistance genes (ARGs) were present in all samples, regardless of grazing treatment; the majority of these encoded efflux pumps and antibiotic modification enzymes (e.g., transferases).
In addition to posting results on the farm website, we shared the outcomes of our study with other farmers and agricultural professionals at the Oregon State University Extension Small Farms Conference in 2021. This project substantially improves our understanding of soil microbial communities under MSRG and provides information for other farmers following sustainable agriculture practices.
Objective 1: Measure the impact of MSRG on soil microbial diversity, composition, and function.
We collected soil samples before, during, and after MSRG and use shotgun metagenomics to identify taxonomic, phylogenetic, and functional characteristics of soil microbial communities (bacteria, fungi, and archaea). We hypothesized that each livestock species has a different effect on soil microbial communities (due to distinct foraging/grazing strategies and unique manure composition), and that MSRG would lead to higher microbial taxonomic and functional diversity, as well as different taxonomic composition, in comparison to monospecies grazing or no livestock.
Objective 2: Assess changes in livestock health and pasture forage quality following MSRG.
Since MSRG can effectively break livestock parasite cycles, we hypothesized that animal health would improve following MSRG. Additionally, we hypothesized that pasture productivity and plant diversity would increase after MSRG, due to changes in manure deposition and grazing patterns.
Objective 3: Share study outcomes with other farmers, agricultural professionals, and the general public.
We disseminated our project methods and results by presenting at the Oregon State University Extension Small Farms School in 2021 and posting results on our farm website.
The major milestones (Figure 3) that are essential to this project are: collecting soil samples and measuring animal/pasture health before (April 14, 2020), during (June 30, 2020), and after (September 16, 2020) MSRG; processing and analyzing microbial samples; presenting the results and methods to other farmers and agricultural professionals at the OSU Small Farms Conference (February 20, 2021); and hosting a Farm Field Day at Black Tansy Farm (May, 2021).
[caption id="attachment_603446" align="aligncenter" width="1024"]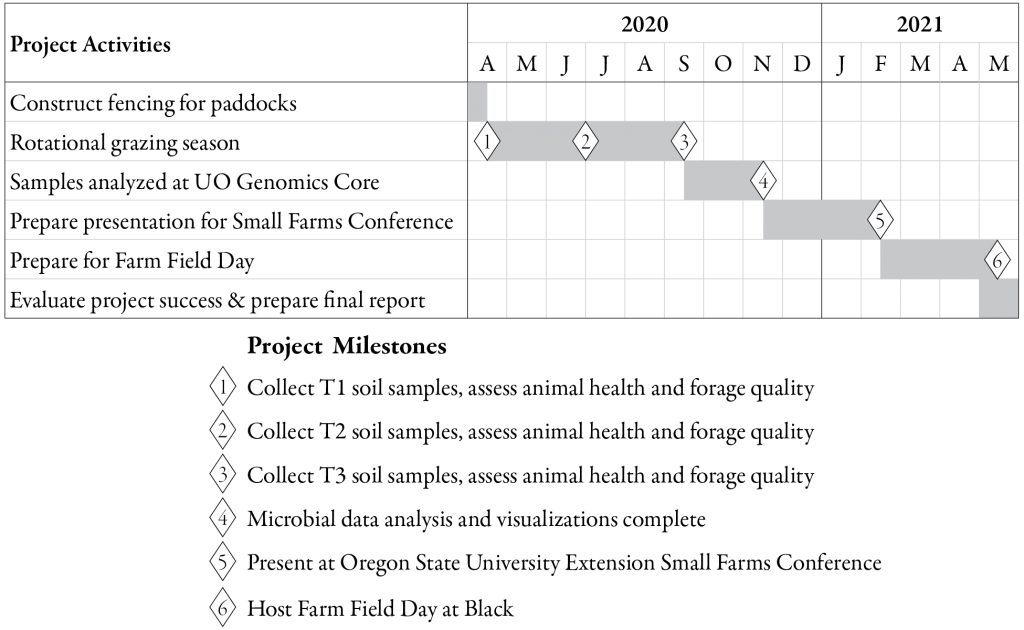 Timeline for project activities and major milestones.[/caption]
Timeline for project activities and major milestones.[/caption]Cooperators
- - Technical Advisor
- - Producer
- (Researcher)
Research
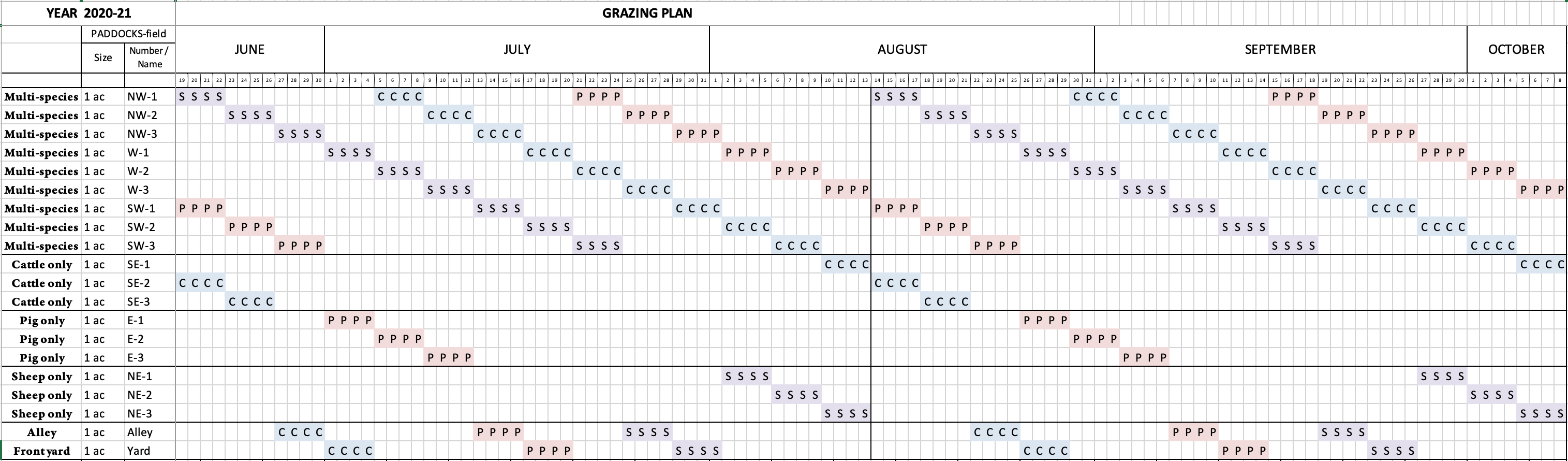
This research was conducted at Black Tansy Farm, a small family-owned and -operated farm located on the outskirts of Springfield, Oregon. The farm was established in 2014 and currently produces meat and fiber from cattle, sheep, and pigs for direct sale to consumers. To achieve the objectives of this project, approximately 2000 feet of new permanent fencing was constructed and additional electric fencing was installed to implement the MSRG system. New fence construction was performed in May, 2020, immediately upon notification of award. As constructed, the ~18 acre pasture was divided into 6 sections, which were further subdivided into 1-acre paddocks with electric fence/netting. The rotational grazing regime began in mid-June, slightly behind the planned timeline due to difficulties scheduling sample collection, and grazing rotations continued until mid-October. In the MSRG plan, sheep went first through each multi-species paddock, as they have the highest nutritional needs, followed by cattle, and then pigs last to break up the soil and manure clods. Instead of performing management-intensive grazing, we elected to follow a strict rotation period of 4 days. The reasoning behind this decision was to ensure sufficient time (46 days) to break the parasite cycle before a given species returned to their first paddock, and to minimize variability in impacts to the soil microbial communities due to differing lengths of time spent on each paddock. See Figure 1 for MSRG plan details.
Objective 1: Measure the impact of MSRG on soil microbial diversity, composition, and function
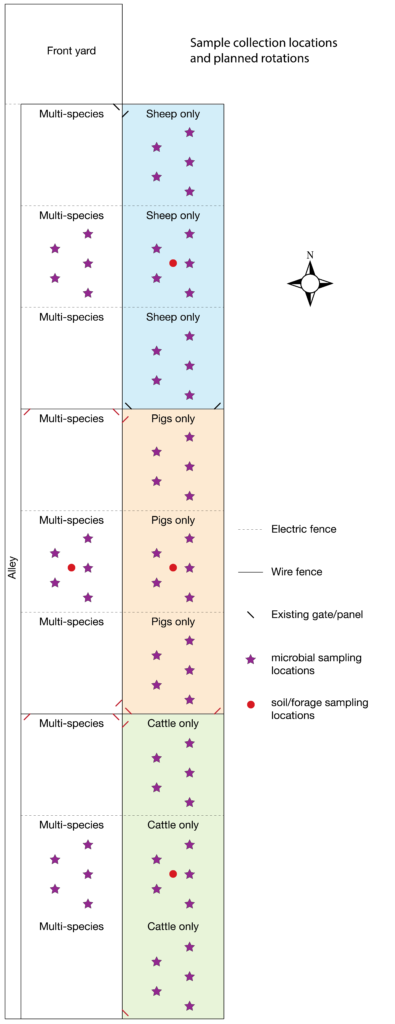
For soil microbial analysis we followed Earth Microbiome Project protocols. Soil cores 2.5 cm in diameter by 10 cm deep were collected from five locations in a roughly "W" shape within selected paddocks (Figure 2) using sterilized stainless steel soil probes. The sample collection paddocks were different from those in our original proposal, because comments from the technical review panel indicated that a balanced number of samples (3 each) representing multi- versus single-species would be a stronger study design than our original plan, which would have resulted in only 1 sample each for cow, pig, and sheep paddocks, but 9 for multi-species. Samples were collected prior to beginning MSRG (mid-June), to provide a baseline condition, halfway through the grazing season (mid-August), and at the end of the grazing season (mid-October) to assess changes in soil microbial community structure and function. For each paddock and time point, the five soil cores were combined and homogenized, then aliquoted into two 0.25 gram subsamples; the remainder of the bulk soil was discarded, except for the final October samples, which were delivered to the local Natural Resources Conservation Service (NRCS) office for soil physical and chemical analysis. From the two technical replicates aliquoted from each sample, we extracted metagenomic DNA using the MoBio PowerSoil DNA extraction kit and prepared libraries for shotgun metagenomic analysis using the Illumina Nextera XT DNA library prep kit. Negative and positive controls were included for quality control, and samples were randomized during extraction and DNA preparation to minimize batch effects. A total of 72 soil samples were submitted on December 10, 2020, for sequencing on the Illumina HiSeq4000 platform at University of Oregon's Genomic & Cell Characterization Core Facility. Raw data was received back from the Core on March 8, 2021, but data cleaning and analyses have not been performed yet.
Objective 2: Assess changes in livestock health and pasture forage quality following MSRG
Body condition scores were tracked from beginning to end of the grazing season to assess any impact of MSRG on livestock productivity. We also recorded disease symptoms and mortalities that occurred during the grazing season. To evaluate how MSRG affected forage quality, we measured forage biomass production by hand clipping samples, and recorded plant species composition in 12" squares at 3 locations (representing low, medium, and high production) for each paddock immediately prior to the final set of rotations. The original proposal also included counting grass tillers in the 12" squares, but this was quickly found to be infeasible. Hand-clipped samples were oven-dried at 140 ºF and weighed; the average of the dry weights were used to estimate forage production for each paddock following the methods in Shewmaker and Bohle (2010).
Homogenized bulk soil remaining from the final samples collected in October were also tested for pH, bulk density, percentage organic matter, plant available nitrogen (N), exchangeable cations (P, Ca, Mg, K), and micronutrients (Zn, Mn, Cu, Fe, B) at AgSource Laboratories in Junction City, Oregon. We had originally planned to send samples to Oregon State University's (OSU) Central Analytical Lab, but chose to use the same lab as our 2019 soil analyses since we learned that different labs may generate different results.
We used shotgun metagenomic sequencing to characterize soil microbial community taxonomy and antibiotic resistome. Results demonstrated broad microbial diversity in all paddock soil microbiomes. Samples collected early in the season tended to have greater archaeal and bacterial alpha diversity than samples collected later for all grazing treatments, while no effect was observed for fungi or viruses. Beta diversity, however, was strongly influenced by both grazing treatment and month for all microbial kingdoms, suggesting a pronounced effect of different livestock on microbial composition. Cattle-only and swine-only paddocks were more dissimilar from multi-species paddocks than those grazed by sheep. We identified a large number of differentially abundant taxa driving community dissimilarities, including Methanosarcina spp., Candidatus Nitrocosmicus oleophilus, Streptomyces spp., Pyricularia spp., Fusarium spp., and Tunggulvirus Pseudomonas virus phi-2. In addition, a wide variety of antibiotic resistance genes (ARGs) were present in all samples, regardless of grazing treatment; the majority of these encoded efflux pumps and antibiotic modification enzymes (e.g., transferases).
Despite the importance of microbial-mediated processes to agricultural productivity and environmental quality, using microbial indicators of soil health remains fraught with technical and conceptual challenges, such as lack of connection to actionable management practices and context-dependent definitions of “good” and “bad” soil microbes. This study contributes valuable knowledge about soil microbial communities in grazing lands, which will help build the knowledge base needed to make future microbial assessments more actionable. Nonetheless, there were several limitations to this work.
This was a small-scale, farmer-initiated study encompassing only a single grazing season. Thus, effects of grazing management must be interpreted with caution, since soil microbial communities can be dynamic from year to year. Additionally, we used shallow sequencing methodology to achieve broad characterization of the most abundant taxa from each microbial kingdom, being mindful that many rare taxa and antibiotic resistance genes would not be recovered. Deeper sequencing in future studies could reveal further differentiation of community structure and antibiotic resistom. Lastly, absolute abundance cannot be inferred from our data; future farm soil studies using quantitative measurement methods would be valuable. This highlights some of the challenges involved with conducting rigorous empirical research in the context of an active commercial livestock operation and suggests future directions to extend this study.
Research Outcomes
Education and Outreach
Participation Summary:
Objective 3: Share study outcomes with other farmers, agricultural professionals, and the general public
We shared our study results with farmers and agricultural professionals at the OSU Small Farms School in September, 2021, and shared our experiences with grant proposal writing with other farmers at the OSU Small Farms Conference in February, 2022. Due to continuing impacts of the COVID-19 pandemic, we were unable to host the planned Farm Field Day, although we did host a farm tour for 12 interns from Deck Family Farm in June, 2021. We also published the study results in the special collection Livestock Production and the Functioning of Agricultural Ecosystems: Volume II of the peer-reviewed journal Frontiers in Sustainable Food Systems. Finally, we have posted our study outcomes on the Black Tansy Farm website (https://blacktansyfarm.com/research/) for the general public to access.