Final report for GNC16-231
Project Information
Cover crop adoption in the Upper Mississippi Basin has increased by 212% over the last 5 years, and farmers have a working understanding of how cover crops can be used as tool to increase soil health and reduce nitrate loading from tile-drained fields to surface waters. However, little is understood about how the inclusion of cover crops influences the soil microbial community, their efficiency in facilitating nitrogen (N) release from cover crop residue, and the relationship between the soil microbiome and cover crop carbon (C) and N release over the cover crop decomposition period. Therefore, the proposed objectives of this study are to i.) Determine the impact of cover crop species and reduced tillage treatments on soil inorganic N following cover crop termination in a corn cropping system, ii.) Evaluate the impact of cover crop species and reduced tillage treatments on soil enzyme activity dynamics as it relates to N mineralization among treatments, and iii.) Evaluate the impact of cover crop species and reduced tillage treatments on the short term dynamics of the soil microbiome during winter cover crop decomposition. The research treatments were established in 2015, thus our analysis will consist of a one-year effect. Systematic soil sampling occurred post cover crop termination during the spring and summer, which detected changes in soil inorganic N concentrations and the facilitating soil microbial community over time. Results indicate that soil β-glucosidase activity at the V6 growth stage was significantly greater for cereal rye dominated treatments relative to hairy vetch treatments, but was similar to the control (p < 0.05). This elevated soil β-glucosidase activity was related to a subsequent increase in soil inorganic N at the following sample date. Soil inorganic N trended higher from the V6 to R2 growth stages in the spring tillage cover crop treatments compared to the spring tillage no cover crop control. Permutational multivariate analysis of variance (PerMANOVA) of phylogenetic based distance matrices revealed that the main effects of cover crop treatment, tillage system, and sampling date (p < 0.05) were significant determinants of soil microbial community composition over the decomposition period.
Description
The overarching goal of this study is to develop a comprehensive understanding of how cover crop species, reduced tillage treatments, and soil microbial community diversity are related to soil inorganic nitrogen availability and cover crop carbon release over the cover crop decomposition period.
Objectives
- Determine the impact of cover crop species and reduced tillage treatments on soil inorganic N following cover crop termination in a corn cropping system
- Evaluate the impact of cover crop species and reduced tillage treatments on soil enzyme activity dynamics as it relates to N mineralization among treatments
- Evaluate the impact of cover crop species and reduced tillage treatments on the short term dynamics of the soil microbiome during winter cover crop decomposition
Cooperators
- (Researcher)
- (Researcher)
Research
Experimental Design
The principle strategy of the proposed research approach was to use systematic soil samplings to evaluate the return of N and C from cover crop residue to the soil ecosystem with relation to soil enzyme activity and the soil microbial community structure. This macro-analysis of the change in soil inorganic N concentrations with time and a molecular analysis of the change in the soil microbial community was examined. The experimental site of the proposed study was located at the Purdue Agriculture Research and Education (ACRE) farm in West Lafayette, Indiana on a silty clay loam soil. The experimental design was split-plot with cover crop as the main plot and tillage as the sub-plot with three field replications (block). Cover crop treatments included (Control, Cereal Rye, Hairy Vetch, Cereal Rye/Hairy Vetch mixture). Hairy vetch (legume) and Cereal rye (monocot) cover crops were specifically chosen because of their physiological and biochemical differences, different methods of scavenging N, their ability to grow in cold weather, their potential to contain distinctly different microbial communities within their surrounding soil microbiomes, and the increasing trend for these species to be used in the North Central Region. Treatments were imposed on field-scale research plots. Each plot was 12 rows wide by approximately 80 m in length. Cover crops were planted on September 28-30, 2015. The cover crops grew over the winter and were chemically terminated on April 14, 2016. Following termination, anhydrous ammonia was applied at a rate of 78 kg ha-1 of N. On April 25, 2016, the spring tillage plots were disk tilled twice to a depth of approximately 15 cm. On April 26, 2016, the same plots were cultivated to prepare the seedbed. The no tillage plots were not disk tilled. On April 26, 2016 maize (Zea mays) was planted in all plots. The research treatments were established in 2015, thus our analysis will consist of a one-year cover crop effect. Prior to these cover crop and tillage regimes, the site was managed in a corn-soybean annual rotation with spring tillage prior to corn planting every other growing season.
Soil Sampling
In the spring, soil sampling at the depth of 0-4 cm was conducted in each plot for each treatment. At each soil sampling date, soil temperature and gravimetric water content was determined. To capture the heterogeneity across plots, 15 soil samples were randomly collected within plots, homogenized. These homogenized samples were then separated into sub-samples. One sub-sample of soil was collected for inorganic N (NO3-N and NH4-N) analysis, and these samples were air dried, ground, sieved, extracted, and analyzed colorimetrically using the SEAL AQ2 analyzer. A second sub-sample was used to determine gravimetric water content by oven drying at 105 degrees Celsius for 48 hours. An additional sub-sample was stored at 4°C, for no longer than 48 hours, and then used to determine soil β-glucosidase (1) enzyme activity. A sub-sample was stored at -20°C for DNA extraction. Soil samples were taken during the cover crop decomposition period to track the cover crop and tillage effect on enzyme activity and the soil microbiome at the VE, V3, V6, VT and R2 corn growth development stages. Considering the treatments are replicated three times, this led to 120 total soil samples for DNA sequencing. Additional soil samples were collected prior to corn planting (April 17th) and at corn physiological maturity (R6) on September 23rd for urease enzyme activity (2) and soil inorganic N analyses. Statistical analysis of soil β-glucosidase activity was completed using the PROC MIXED procedure in SAS (3).
Molecular Microbial Ecology Methods
Soils were sampled for DNA sequencing on five sampling dates. Soil microbial communities (Bacteria) were characterized using the small subunit (16S) rRNA gene sequences determined using the Illumina MiSeq system. Total genomic DNA was extracted from soil using the QBioGene soil kit that has been successfully used in the Nakatsu laboratory at Purdue (4, 5). Primers that target the 16S rRNA genes of Bacteria were used for PCR. Primers will have duel index tags to differentiate multiple samples in a single run (6). PCR was carried out using Q5® High Fidelity DNA Polymerase (New England Biolabs) to minimize error rates (100 times lower than Taq polymerase). PCR amplicons will be purified using Agencourt AMPure XP kit (Beckman) and quantified by fluorometry after staining using the QuantiFluor dsDNA System. Amplicons from each sample were combined in equivalent quantities and sent to the Purdue Genomics core facilities for 2×250 paired end sequencing using the MiSeq Illumina instrument. Sequence reads were pre-processed to remove primer/tags and low quality sequences then pair ends were merged. Sequences were analyzed using the QIIME pipeline (7) and the vegan package (8) in R version 3.3.2 (9). The sequencing and sequence analysis methods (including diversity measures) previously optimized and successfully used for other projects in the Nakatsu laboratory (10) were used in this project.
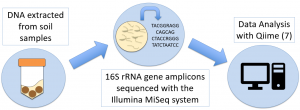
Figure 1: Summary of molecular microbial ecology methods.
References
- Eivazi, F., Tabatabai, M.A. 1988. Glucosidases and galactosidases in soils. Soil Biol. Biochem. 20, 601–606.
- Kandeler, E. & Gerber, H. 1988. Short term assay of soil urease activity using colorimetric determination of ammoniuim. Biol Fert Soils. 6: 68.
-
SAS Institute Inc., SAS Campus Drive, Cary, North Carolina 27513.
- Joynt J., M. Bischoff, R.F. Turco, A. Konopka, C.H. Nakatsu. 2006. Microbial community analysis of soils contaminated with lead, chromium and organic solvents. Microb. Ecol. 51:209-219.
- Sundberg C., W.A. Al-Soud, M. Larsson, E. Alm, S.S. Yekta, B.H. 2013. Svensson, . . . A. Karlsson. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol. Ecol. 85:612-626.
- Gloor G.B., R. Hummelen, J.M. Macklaim, R.J. Dickson, A.D. Fernandes, R. MacPhee, and G. Reid. 2010. Microbiome profiling by Illumina sequencing of combinatorial sequence-tagged PCR products. PLoS ONE 5:e15406.
- Caporaso J.G., J. Kuczynski, J. Stombaugh, K. Bittinger, F.D. Bushman, E.K. Costello,. and R. Knight. 2010.QIIME allows analysis of high-throughput community sequencing data. Nat. Meth. 7:335-336.
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2015) Vegan: community ecology package. R package version 2.2–1 available at https://cran.r-project.org/web/packages/vegan/index.html
- R Core Team (2014) R: a language and environment for statistical computing. R version 3.3.2 available at https://cran.r-project.org/bin/windows/base/old/3.3.2
- Nakatsu C.H., A. Armstrong, A.C. Clavijo, B.R. Martin, S. Barnes, and C.M. Weaver. 2014. Fecal bacterial community changes associated with isoflavone metabolites in postmenopausal women after soy bar consumption. PLoS ONE.
Research Results and Discussion
Overall, this data is preliminary and should not be used, recreated, or presented outside of this report. This NCR-SARE grant funded one summer of a multi-summer study, and this allowed the researchers to better understand the seasonal trends of soil enzyme activity and inorganic N availability.
Objective 1: Determine the impact of cover crop treatment and tillage treatments on soil inorganic N availability following cover crop termination during the decomposition period
Soil inorganic N (mg of inorganic N per kg soil) was measured at seven sampling dates during the corn growing season following cover crop termination (Figure 2). Cover crop treatment did not have a significant effect on soil inorganic N availability, unlike tillage system (Table 1). Soil inorganic N peaked from the VT-R2 corn growth stages (June 28th-August 1st). During this peak, soil inorganic N trended greater in the spring tillage cover crop treatments compared to the no-tillage treatments. At VT, when soil inorganic N was greatest in the spring tillage treatments, the cover crop treatments trended higher than the no cover crop control.
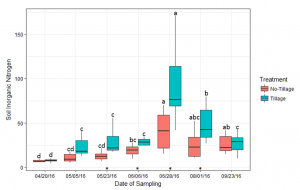
Figure 2: Average soil inorganic nitrogen (mg Kg-1) for tillage and no-tillage residue management systems during the corn growing season. Bars with the same letter are not statistically significant from each other within tillage treatment over the growing season (p < 0.05). Significant differences within each sampling date are signified by an asterisk (*) directly above the sampling date on the x-axis (p < 0.05)
Table 1: ANOVA Table detailing effect of cover crop, tillage, and sampling date on inorganic N availability
|
Table 1. Inorganic Nitrogen ANOVA Table |
||
|
|
2016 |
2017 |
|
Date |
<0.001 |
<0.001 |
|
Cover Crop |
NS |
NS |
|
Tillage |
<0.001 |
NS |
|
Date*Cover Crop |
NS |
NS |
|
Cover Crop*Tillage |
NS |
NS |
|
Date*Tillage |
<0.001 |
0.04 |
|
Cover Crop*Tillage*Date |
NS |
NS |
Objective 2: Evaluate the impact of cover crop treatments and tillage systems on soil enzyme activity dynamics as it relates to N mineralization among treatments.
Soil enzyme activity was measured at seven sampling dates over the cover crop decomposition period. Enzyme activity assessments included the carbon cycling enzyme β-glucosidase and N cycling enzyme urease. Soil β-glucosidase activity was elevated on June 6 at the corn V6 growth stage (Figure 3). This was one sampling date prior to the spike in soil inorganic N (Figure 2). On June 6, soil β-glucosidase activity in the cereal rye-based treatments was significantly greater than the hairy vetch treatment, but was similar to the control (p < 0.05). Soil β-glucosidase activity in the cereal rye-based treatments was significantly greater on June 6 compared to the other seven sampling dates in all treatments but HV, which had similar activity levels on May 23.
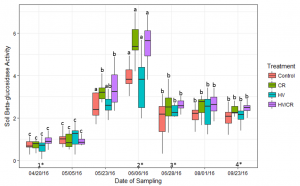
Figure 3: Measured average levels of soil β-glucosidase activity (μmoles p-nitrophenol released g dry soil-1 hr-1) in each cover crop treatment during the 2016 corn growing season. Bars with the same letter are not statistically significant from each other within cover crop treatment over each corn growing season (p < 0.05). There was a significant difference between cover crop treatments within four sampling dates (p < 0.05) (1*,2*,3*,4*). (1*) The HV/CR treatment was significantly different from the HV treatment. (2*) The cereal rye based treatments were significantly different from the HV and Control treatments. (3*) The HV/CR treatment was significantly different from the Control treatment. (4*) The cereal rye based treatments were significantly different from the Control, and the HV/CR treatment was significantly different from the HV.
Soil urease activity increased with soil temperature until the corn R2 growth stage on August 1 (Figure 4). On May 23 (V3) urease activity in the hairy vetch treatment was significantly less than the cereal rye treatments (p < 0.05).
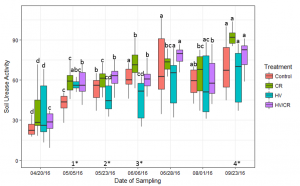
Figure 4: Measured average levels of soil urease activity (μg NH4-N g dry soil-1 2 hr-1) in each cover crop treatment during the 2016 corn growing season. Bars with the same letter are not statistically significant from each other within cover crop treatment over each corn growing season (p < 0.05). There was a significant difference between cover crop treatments within four of the sampling dates (p < 0.05) (1*,2*,3*,4*). (1*) The CR, HV/CR, and HV treatments were significantly different from the control. (2*) The cereal rye based treatments were significantly different from the HV treatment. (3*) The HV treatment was significantly different from the other three treatments, and the CR was significantly different from the HV/CR. (4*) The cereal rye treatment was significantly different from the HV and Control treatment.
Objective 3: Evaluate the impact of cover crop species and reduced tillage systems on the short term dynamics of the soil microbiome during winter cover crop decomposition.
Following sequencing, a cutoff of 85,802 reads was used for analysis and all 120 samples were included. The Good’s Coverage ranged from 90-92% for the samples (average 91%). The main effect of no-tillage residue management treatment had a significantly greater Shannon Diversity Index than the spring tillage system (not shown). Also, the Shannon Diversity Index was significantly lower at the beginning of the cover crop decomposition period on May 5th compared to the following four sampling days (not shown). This could be due to the lack of cover crop decomposition early in the decomposition period at the beginning of May and then more rapid decomposition as soil temperatures increased later in the corn-growing season.
The main effects of cover crop treatment, tillage system, and sampling date were all significant determinants of soil microbial community composition. A weighted normalized Unifrac distance matrix was created between samples through a beta diversity analysis to examine phylogenetic metrics. Permutational multivariate analysis of variance (PerMANOVA) with 999 permuations of the distance matrices revealed the significance of the main effects. Principle coordinate analyses (PCoA) were conducted using these distance matrices to observe the separation of samples by the main effects. The main effect of cover crop treatment was a significant determinant of the soil microbial community composition (p < 0.001) as illustrated in the separation of the samples in the PCoA below (Figure 5). Likewise, the main effect of tillage system was significant (p < 0.003) (Figure 6) as well as sampling date (p < 0.001) (Figure 7). In addition, to determine if there was a cover crop effect all cover crop treatments were grouped and compared against the control treatments (Figure 8). The PerMANOVA of the distance matrix revealed that there was a significant difference between plots that were cover cropped and the control plots (p < 0.001).
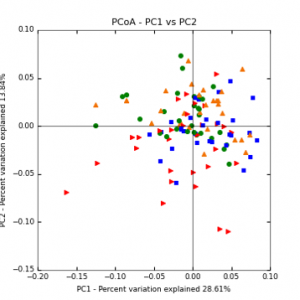
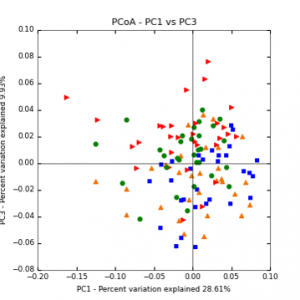
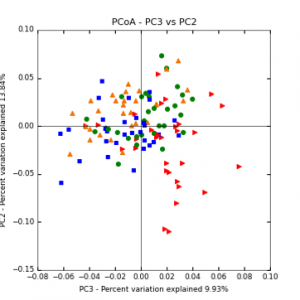
Figure 5: PCoA plot of weighted_normalized_Unifrac distance matrix illustrating groupings of samples by cover crop treatments
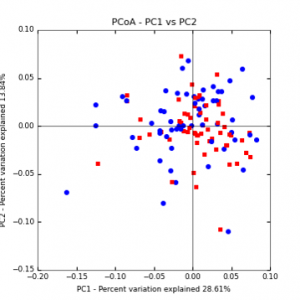
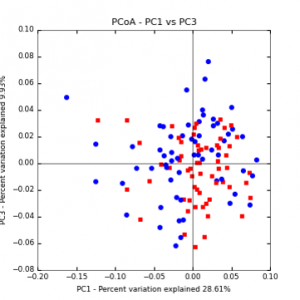
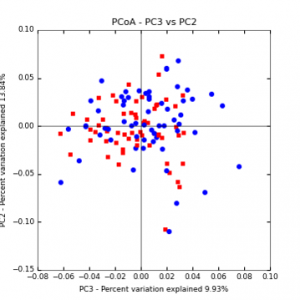

Figure 6: PCoA plot of weighted_normalized_Unifrac distance matrix illustrating groupings of samples by tillage treatment
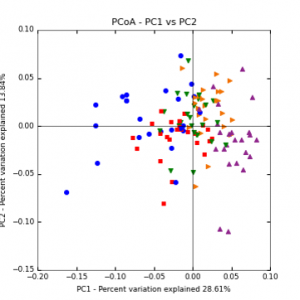
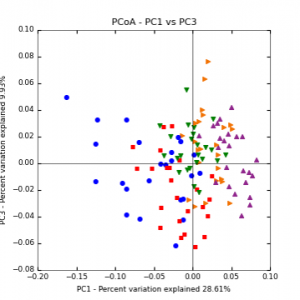
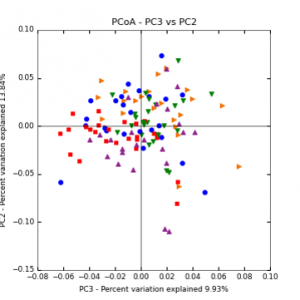

Figure 7: PCoA plot of weighted_normalized_Unifrac distance matrix illustrating groupings of samples by sampling date
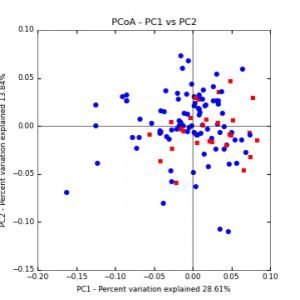
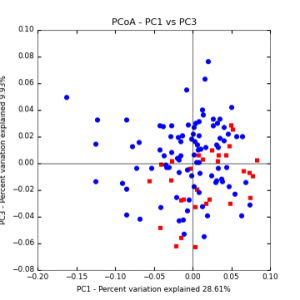
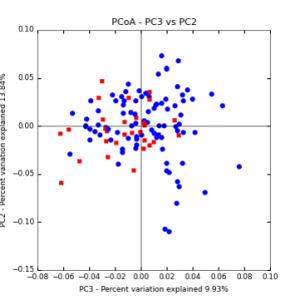

Figure 8: PCoA plot of weighted_normalized_Unifrac distance matrix illustrating groupings of samples by cover crop treatments compared to control treatments
After determining that the main effects of cover crop treatment, tillage treatment, and sampling date were significant we investigated the impact of cover crop treatments within each of the sampling dates. Table 2 illustrates that there was a cover crop effect within each of the sampling dates. The effect of cover crops on May 5 was marginally significant (p < 0.1), and the effect at the following for sampling dates was significant (p < 0.05).
Table 2: Effect of cover crop treatment within each of the sampling dates during the cover crop decomposition period
| Sampling Date | PerMANOVA | |
| Test Statistic | P-value | |
| May 5th | 1.45 | 0.092 |
| May 23rd | 1.77 | 0.012 |
| June 6th | 1.61 | 0.021 |
| June 28th | 1.54 | 0.041 |
| August 1st | 2.04 | 0.008 |
Educational & Outreach Activities
Participation Summary:
The results of this research project were disseminated to farmers and agriculture professionals as a poster or oral presentation by Clayton Nevins at educational events (detailed below). These events occurred at scientific conferences, including the 2016 and 2017 Midwest Cover Crop Council Meetings. Other events include the Graduate Student Plant Science Symposium and Indiana Corn Marketing Council Corn Showcase at Purdue University. Dr. Shalamar Armstrong has presented the results of this project at many presentations to farmers, certified crop advisers, and agriculture professionals at several events during the duration of the project. The results of this research will continue to be presented in a manner that reaches farmers and agriculture professionals by the Armstrong Research Group at Purdue University at the conclusion of this project. In addition, the authors aim for this project to result in a peer-reviewed journal article publication. Now, as we finish the data analysis of this study, the manuscript for this work is in preparation.
Nevins, C.J., C. Lacey, L. Hoagland, R. Turco, C. Nakatsu and S. Armstrong. October 2017. Soil microbial community dynamics over the growing season of a corn agroecosystem after winter cover cropping. Oral presentation at the Agronomy, Crop and Soil Science Societies 2017 Annual Meeting. Tampa, Florida
Nevins, C.J., C. Lacey, L. Hoagland, R. Turco, C. Nakatsu and S. Armstrong. October 2017. Impact of cover crop species on enzyme activity and nitrogen supply at corn critical growth stages. Poster presented at the Agronomy, Crop and Soil Science Societies 2017 Annual Meeting. Tampa, Florida
Nevins, C.J., C. Lacey, L. Hoagland, R. Turco, C. Nakatsu, and S. Armstrong. August 2017. Soil microbial community dynamics over the growing season of a corn agroecosystem after winter cover cropping. Poster presented at the Purdue-Pioneer Graduate Student Plant Science Symposium. West Lafayette, Indiana.
Nevins, C.J., C. Lacey, L. Hoagland, R. Turco, C. Nakatsu and S. Armstrong. March 2017. Influence of cover crop species on enzyme activity and nitrogen supply at corn growth stages. Poster presented at the Midwest Cover Crop Council Annual Meeting. Grand Rapids, Michigan.
Nevins, C.J., C. Lacey, L. Hoagland, R. Turco, C. Nakatsu, and S. Armstrong. November 2016. Influence of cover crop species on enzyme activity and nitrogen supply at critical corn growth stages. Poster presented at the Agronomy, Crop and Soil Science Societies 2016 Annual Meeting. Phoenix, Arizona.
Project Outcomes
As we learn more about the soil microbiome and the ecosystem services carried out by the microbiome, we increase our understanding of the timing of nutrient release and availability in these systems. Enhancing our understanding of these systems could lead to a decrease in the reliance on synthetic fertilizer. This decrease in synthetic inputs and increase in knowledge about winter cover cropped agroecosystems is economically attractive to the North Central Region farming community and can result in increased environmental sustainability and agriculture productivity.
With a dearth of literature available focused on the investigation of cover crop systems using next-generation sequencing technologies there was a lot to gain from this research. From this study, we now know that the soil microbiome is impacted by the species of winter cover crops that farmers in the Midwest Corn Belt routinely use in their sustainable management plans. This research also gave us insight into the bacteria abundance patterns in agroecosystems with different cover crops and tillage treatments. Overall, there were three key findings from this research: 1) Cover crops impact the soil microbiome at each sampling date over the decomposition period, 2) Tillage system impacted the soil microbial community over the decomposition period, and the impact was greatest over a month after cover crop termination on May 23, and 3) There is a relationship between soil β-glucosidase activity and inorganic N availability, that had not been previously reported in the scientific literature.
Gaining this knowledge has made the researchers in the Armstrong Lab at Purdue more ambitious to develop a more comprehensive understanding of the short-term dynamics of the soil microbiome and nutrient cycling in winter cover cropped sustainable agroecosystems. In addition, while presenting the findings of this research it is enlightening to hear from farmers and agriculture professionals interested in the findings of this research. This led to an increase in awareness of the issues that are at the forefront of the pursuit to make agriculture more sustainable in the North Central Region, which the Armstrong Lab believes is the cover cropped agroecosystems.
Recent advancements in high-throughput sequencing technology has made it feasible to generate large amount of sequencing data in a cost and time efficient manner. Investing in research that utilizes molecular microbial ecology techniques to better understand the soil microbial community abundance patterns and plant-soil-microbe interactions could positively influence farmers’ investments in sustainable agriculture techniques such as cover cropping and no-tillage.