Final report for GNE17-143
Project Information
Objective 1:
Use marker-assisted selection (MAS) and background genome selection to rapidly transfer BLS resistances (Rx-4 and QTL11; on chromosome 11) into well-adapted fresh market tomato breeding lines that already possess strong tolerance to EB (EBT) and resistances and LB (Ph-2 and Ph-3). Outcome: This is the first step to bringing BLS resistance into fungal resistant tomatoes. This Rx-4 and QTL11 “coupling phase” linkage provides broad resistance to BLS, but full control requires Rx-3 (on chromosome 5), which is added in objective 2.
Objective 2:
Use marker-assisted selection and background genome selection to rapidly transfer the BSk (Pto), BLS (Rx-3), and SLS (SLS-2) linked resistances on chromosome 5 into the same EB, LB, and BLS resistant tomato being produced in objective 1. Outcome: The combination of Rx-4, QTL-11, and Rx-3 provides the best resistance to all four species of Xanthamonas infecting tomato. The addition of Pto provides a valuable resistance to the primary race of Pseudomonas, race 0, causing BSk. The addition of Pto resistance to bacterial speck and the strongest current resistance to bacterial spot, in lines with genetic control of the major fungal/oomycete diseases of Northeast tomato, would provide an unprecedented level of disease control in fresh market tomato.
Joint outcome:
The combined results of objectives 1 and 2 will also produce a set of near-isogenic lines (NILs) containing different subsets of bacterial and fungal resistance genes/QTL in adapted fresh market tomato. These are valuable in breaking linkage drag, if present, and to plant pathologists studying interactions among resistance genes and pathogens.
Objective 3:
A) Field trial the lines and isolines created by objectives 1 and 2 to determine the acceptability of their horticultural plant/fruit type under low disease control input conditions, permitting the observation of natural infection. B) Also, perform tests of lines with/without BLS resistances (Rx-3 and QTL-11/Rx-4) to determine the degree of control possible in fresh market lines. Outcomes: Field trials will determine the impact of the chromosome 5 and 11 introgressions containing the bacterial resistance genes on the horticultural characteristics of the resulting lines. Characteristics include plant type, fruit size and quality, productivity and maturity. This will determine whether the transfer of either introgression also carries deleterious traits to be eliminated. BLS control will be assessed through naturally and artificially infected replicated trial plots in Cooperation with collaborating breeders and extension/plant pathologists over two years.
Cooperators
Research
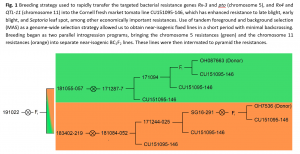
Marker-assisted backcross breeding scheme: A Cornell fresh market line, CU151095-146, with strong resistance to late blight, early blight, and Septoria leaf spot was crossed to two donor parents from the Francis OSU breeding program that bore distinct resistance packages. These packages were initially brought into the Cornell genetic background independently but are now unified in a small number of final lines. A schematic of the breeding scheme pedigree is shown in Figure 1. The first Ohio processing tomato line, OH7536, possesses a coupling phase linkage between the Pto and Rx-3 bacterial resistance genes on chromosome 5. The second, OSU line OH087663, possess a coupling phase linkage between two targeted bacterial spot resistance genes, QTL-11 and Rx-4, on chromosome 11.
Tandem foreground and background marker-assisted selection (MAS) was used to rapidly transfer and combine these resistances into one of our best fresh market tomato backgrounds. Solcap SNPs were adapted into PCR, KASP, and amplicon sequencing-based markers for use in positive foreground selection for targeted regions encoding bacterial resistance genes, and for negative background selection against donor genotypes in the non-target genome to rapidly recover the recurrent parent genetic background. In each segregating generation, MAS was performed in two phases to minimize costs associated with genotyping and greenhouse space and maximize time use-efficiency. Foreground selection was performed on a large population using only a few genetic markers, followed by background selection on the selected subset of the population using genome-wide SNPs placed evenly across a high-density genetic distance map. A small number of selected plants, typically 3-5, were then used to establish the following generation.
Transfer of Bacterial Spot Resistances on Chromosome 11.
BC1F1: After analysis of a BC1F1 population (n = 285), 3 plants were identified that possessed the full chromosome 11 region but were otherwise were 80+% identical by descent to the recurrent fresh market parent. This degree of progress is what would be expected for two cycles of backcrossing without background selection. The selected BC1F1 plants were backcrossed to the fresh market parent to produce BC2F1 seed.
BC2F1: When this strategy was repeated summer 2017 with a BC2F2 population (n = 192), 10 plants were identified that were heterozygous for the full chromosome 11 region and were otherwise 98+% identical by descent to the recurrent parent. The selected BC2F1 plants were self-pollinated to produce BC2F2 seed lots.
BC2F2: Screening of two BC2F2 populations in the fall of 2017 with PCR markers led to selection of 3 BC2F2 plants that were homozygous for the chromosome 11 region containing the bacterial spot gene Rx-4 and QTL-11 but had otherwise recovered all of the recurrent parent genetic content on all other chromosomes. Additional plants recombinant for the chromosome 11 region were also saved, in case we later discover linkage drag issues associated with the chromosome 11 region. An additional selection was retained that contained an extraneous donor region on the bottom of chromosome 2 that resulted in smaller fruit size but increased overall productivity and the percentage of fruit which were of marketable quality. While not useful for breeding of large-fruited fresh market tomatoes, the lineage derived from this selection is potentially useful for smaller-fruited cherry or plum tomato cultivars.
BC2F3: These BC2F2 selections were self-pollinated in the greenhouse and produced fixed BC2F3 seed, harvested in March 2018. This seed established the new lines combining the fungal resistances of the CU parental line with the Rx-4 and QTL-11 bacterial speck resistances on chromosome 11.
Additional denser genotyping was performed on the BC2F2 lines during the summer of 2018 to confirm that they were homozygous for fresh market parental DNA throughout the genome and to identify any residual heterozygosity.
Transfer of Bacterial Spot and Speck Resistances on Chromosome 5.
Screening of a BC1F1 population (n = 713) for the chromosome 5 transfer was completed in June 2017 resulting in a selection of 4 plants that were heterozygous for the full chromosome 5 region carrying prf /pto and Rx-3 and were heterozygous for 3 or 4 additional regions on other chromosomes. These BC1F1 plants were crossed to the recurrent parent to generate BC2F1 seed.
A BC2F1 population (n = 711) was sown Dec 2017 and screened with chromosome 5 markers in early Jan 2018, identifying 94 plants that were heterozygous for the entire chromosome 5 region carrying both Prf/Pto and Rx-3, and in which heterozygosity for at least one other chromosome was eliminated. Further SNPs on these plants facilitated the identification of 25 plants which were self-pollinated. Several of these BC2F2 plants revealed novel recombinations within the chromosome 5 region, and so these plants were grown to maturity and seed was collected in case later trials indicated a need to reduce the introgression size/break linkage drag.
Three BC2F2 populations were selected for continuation of the project, two of which had three additional segregating regions besides the CH5 introgression, and one of which had just two. Two distinct populations of 384 plants each were planted in Freeville using two of these seed lots during the summer of 2018, while the third was planted on a grower’s field in the Hudson valley and at the Long Island Agricultural Experiment station, in coordination with Dr. Meg McGrath and Teresa Rusinek of Cornell Cooperative extension. PCR markers run on these populations facilitated the identification of several plants which were homozygous for the chromosome 5 region carrying both Pto/Prf and Rx-3 and homozygous for fresh market tomato DNA at all other regions surveyed. Of these, phenotypic observations on important traits such as maturation period, fruit size, set, and shape, and foliar coverage and growth habit was used to identify 7 superior individuals in Freeville, 2 in the Hudson Valley, and 2 in Long Island. Self-pollinated seed was extracted from the fruit of these individuals to establish BC2F3 fixed near-isogenic lines.
Because pto and SLS-2 are very close in location, but in repulsion, there are two types of chromosome 5 lines: those in which the full chromosome 5 introgression was transferred possess pto/Rx3, in the background of CU151095-146, but no longer have SLS-2 for Septoria resistance. Other lines had a recombination reducing the chromosome 5 introgression, and so have Rx3, but not pto, in the background of CU151095-146, that still includes SLS-2. After identifying recombinant individuals using molecular markers in summer 2019 that may bring pto and SLS-2 together in coupling phase, we are now planning a Septoria screen that should indicate whether these two resistances were unified in a single line.
Additional, denser genotyping was also performed on the chromosome 5 lines using semi-random and targeted quantitative sequencing, to determine if they are indeed homozygous for the fresh market parental tomato line throughout the genome, or if any smaller regions of processing tomato DNA remain in the genome that were not previously detected by low density genotyping.
Combining all resistance genes to Bacterial Spot and Speck in fresh market tomato.
Three chromosome 5 lines were crossed to three chromosome 11 lines (in all combinations) to create hybrids to test summer 2019, and to bring together all resistance genes in our fresh market tomato background. F1 seed of this cross was harvested in the greenhouses in Ithaca, NY in fall 2018. Three F1 plants from each cross were then grown up and self-pollinated in winter/spring 2018-2019, producing F2 seed of each group in May 2019, in time for the 2019 field season.
Existing trial data on the parents of each cross were analyzed, allowing us to choose one F2 family that was most likely to have high yield, large fruit size, and an appropriate plant growth habit for the fresh market tomato class. Twelve-hundred individual F2 plants were then grown of from the self progeny of the selected F1 family and screened with PCR markers. Markers flanked the chromosome 5 and chromosome 11 resistance-bearing coupling phase linkages, allowing us to identify 24 plants of each class that were homozygous for all four possible combinations of the two resistance regions (i.e. classes of plants that were homozygous for both the chromosome 5 and chromosome 11 resistances, plants that only had one of these resistance regions, and plants that had completely lost both resistance regions). The 24 plants of each class were then split randomly into four groups of six plants that were planted in a randomized complete block design at the Thompson research farm (Freeville, NY) in summer 2019. This trial allowed us to investigate the impacts of these regions on fruit quality and yield, and plant maturity and growth habit traits. The four classes of plants allowed us to investigate the impacts of the resistances individually and together, with the class without resistances acting as the internal control.
Field trials 2018: During the summer 2019 season, the new lines were tested for plant and fruit characteristics in field trials in three locations in NYS (Long Island, Hudson Valley, Freeville, NY), as well as in several inoculated bacterial spot trials at OSU (Freemont and Wooster). Plant spacings of 3 feet were used in 6 plant plots on black plastic. Fixed CH11 isolines were replicated four times in each trial in New York state. These were uninoculated and primarily focused on identifying and evaluating the effect of the CH11 introgressions on important horticultural traits. OSU trials were on 1ft spacing on bare soils and were inoculated with the pathogen as young transplant starts in the greenhouse.
Field trials 2019: Several finished chromosome 5 and chromosome 11 lines, along with their F1 hybrids and F2 segregating populations were be planted on research plots and farmer’s fields in Freeville NY, Fremont Ohio, Long Island, and the Hudson valley in cooperation with OSU and Cornell Cooperative Extension in the summer of 2019 to evaluate the horticultural performance of these lines and study the effect of the resistances genes (in homozygous and heterozygous conditions) in resistance to bacterial speck and spot diseases under both inoculated and natural infection conditions. Plants were trellised this year in NYS to allow for yield trials in conditions like those on commercial farms. Results from these trials completed the breeding and research goals of this work and generated several breeding lines which can be licensed for use in commercial breeding.
Phenotyping: Phenotyping of the field trials used the following methods. Disease was rated weekly in the inoculated trials on the Horsfall-Barratt scale. Disease data was converted to area under the disease progress curve (AUDPC) for analysis. Maturity and yield were measured by weekly harvests between the last week of August and the first week of October, harvesting all fruit that had reached 50% full color. Harvested fruit were classified as “marketable” or “unmarketable” and weighed and counted to produce measures of marketable fruit size and of primary defects. Fruit were also sliced in half and scanned on a flatbed scanner for use with the tomato fruit analyzer, which provides data on fruit color, shape, uniformity, locule number and size, pericarp thickness and size.
Genetic mapping of Early Blight tolerance in cultivated tomato: In order to effectively pyramid resistances to all five major diseases of tomato using only genetic markers, we needed to map QTL that conferred partial early blight resistance within our genetic population. Early blight resistance is the only disease for which these genetic markers did not yet exist. We planted two populations of ~300 individuals, one an F2 derived from the cross of CU151095-146 and OH087663, and the other a triple cross between F1 created for the aforementioned parents and the OH processing tomato line OH7536, at the Terwilliger research farm in Freeville, NY in the summer of 2017. These were inoculated in the second week of July with an Alternaria tomatophila NC isolate No. 10 spore suspension with a concentration of ~30,000 spore/mL using backpack sprayers and inoculum grown in the laboratory. These were planted in 6-plant plots with 4 ft spacing on black plastic and were irrigated by both drip and overhead irrigation to maintain plant health and encourage the spread of disease. Stem and foliar early blight lesions and blights were rated 3+ times throughout the season, and the results were used to identify plants at the extremes of the disease phenotype distribution that would be selectively genotyped using semi-random and targeted quantitative sequencing. QTL mapping was performed with these selected genotypes (using rQTL) in order to identify the large effect QTL underlying partial EB resistance. Genetic markers (KASP and PCR) were created for highly influential SNP genotypes and used to maintain, and in the case of some of the breeding populations, improve the resistance to EB. Two QTL confirmation populations were grown the following year (summer 2018) to evaluate the heritability of these QTL. These populations included ~17 F2:3 families in a replicated trial and a single segregating population generated from the BC1F2 seed of the chromosome 11 bacterial breeding population. These were planted in 8-plant plots with 2.5 ft spacing on black plastic and were irrigated by both drip and overhead irrigation to maintain plant health and encourage the spread of disease. This work supported the project goal of using marker-assisted selection to pyramid resistances to all five major bacterial, fungal, and Oomycete diseases of northeast-grown tomato into our elite fresh-market background.
Breeding and testing of Bacterial resistances:
Transfer and field testing of Bacterial Spot Resistances on Chromosome 11.
We successfully used tandem foreground and background marker-assisted selection to rapidly introgress the CH11 resistances from the OH parent OH087663 into the background of the CU fresh market tomato line CU151095-146, creating a set of chromosome 11 lines that combine broad spectrum resistances to Xanthamonas sp., narrow-sense resistance to X. perforans (T3), and strong resistances to fungal/oomycete blights including late blight, stem, foliar, and fruit phases of EB, and Septoria leaf spot. We have now performed two years of performance testing of these lines, with uninoculated trials in New York to evaluate horticultural quality/utility, and in inoculated trials in Ohio (in cooperation with Dr. Francis of OSU) to evaluate the disease resistance of the new lines. Denser genotyping on the CH11 materials, performed by semi-random and targeted quantitative sequencing, revealed a small stretch of heterozygosity on chromosome 1 for some lines that did not appear to be correlated with modifications of important performance-related phenotypes.
There have now been two years of field trials of the chromosome 11 introgression lines. A
Marketable yield of the chromosome 11 lines significantly increased over that of the recurrent parent with the introduction of the QTLs on chromosome 11, going from 14.4 Kg of marketable produce per 6-plant plot to 21.5 Kg (p = 0.03), while there was no significant impact of the introgression on average weight of marketable fruit, the percentage of marketable fruit, or days to maturity (defined as number of days from planting until 50% of the fruit are red-ripe). As expected, there was a significant reduction in fruit size for the subset of chromosome 11 lines that carry an extraneous chromosome 2 donor introgression. In all other traits evaluated, such as foliar coverage and uniformity of set and ripening, the chromosome 11 lines performed at least equivalently to the recurrent parent CU151095-146, indicating the successful introduction of these resistance without negatively impacting quality (linkage drag). Chromosome 11 lines also showed a dense plant growth habit with shorter internodes that may be a challenge to field-based growing environments but could be a benefit to high-tunnel production due to the reduction in plant growth area, shorter internodes, and denser fruit clusters.
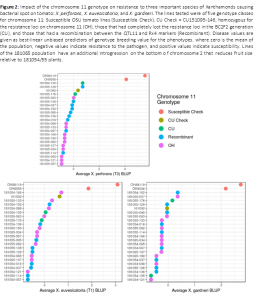
Replicated and inoculated field trials in cooperation with Ohio State University revealed a strong and significant effect (Figure 2); P << 0.001, Type II SS, df = 3,27) of the chromosome 11 introgression from OH7663 at reducing disease symptoms caused by X. perforans (T3). Lines which had functional copies of Rx4 and QTL-11 fared significantly better than the recurrent parent check, susceptible checks, and sister lines that had lost the QTL due to recombination and/or independent assortment. This was as expected, due to the nearly complete control of a true R-gene, characterized by a gene-for-gene interaction between a pathogen effector and a plant pathogen receptor protein. Every line that carried Rx4, and several of the recombinant lines that may have retained Rx4 (The marker density was not high enough to specify where the recombination occurred between the markers flanking the two QTL on chromosome 11 due to a lack of polymorphism between the parents that inhibited the creation of additional descriptive genetic markers), displayed infection severity below the population mean, confirming the successful transfer of an effective bacterial spot resistance gene into the Cornell fresh market tomato background.
Results of the inoculated field trials in Fremont OH were less clear against X. euvesicatoria (T1) and X. gardneri (T2). We had not yet introduced the Rx3 locus (on chromosome 5) into the chromosome 11 lines, so resistance to these pathogens would have been derived from the partial resistance conferred by QTL-11 and any marginal effect of the Rx4 gene against non-target pathogen races. Materials that were homozygous for QTL-11 were in fact significantly more resistant (P<0.05, Type II SS, df = 3,27) to X. euvesicatoria than the Cornell recurrent parent, but curiously were not significantly different from two sister lines that had apparently lost the QTL-11 during recombination and/or independent assortment. This could be due to the small sample size (there were only two of these lines entered into the replicated trial). Alternatively, this could occur from the not-unlikely scenario that the QTL was present in these lines despite the genotypes indicated by the flanking markers. Due to our poor knowledge regarding the exact location of this centromeric QTL, and to the absence of nearby and tightly-linked flanking markers because of low polymorphism between the OH and CU parents on chromosome 11, we cannot rule out this possibility. Indeed, these are the challenges of working within elite, cultivated materials that already have high levels of quantitative resistance and that lack genetic polymorphism due to the significant genetic bottleneck that occurred during domestication.
Finally, there was no significant difference among the genotype classes at the 5% Type I error rate for resistance to X. garneri (P = 0.463, Type II SS, df = 3,27). Reasons for this result may include the relatively high level of quantitative resistance already present in the Cornell materials, the lower trait heritability due to the QTL, or the relatively low disease pressure, and hence smaller phenotypic range, in the X. garneri trial.
Transfer of Bacterial Spot and Speck Resistances on Chromosome 5
We successfully transferred the chromosome 5 resistances from OH7536 into the elite fresh-market tomato line CU151095-146. This creating a set of chromosome 5 lines that combine X. euvesicatoria (T1) bacterial spot resistance from the Rx-3 locus, pto-derived resistance to Pseudomonas syringae p.v. tomato, the causative agent of bacterial speck of tomato, and the previously outlined fungal and oomycete resistances in CU151095-146. There are two types of chromosome 5 lines: those with the full chromosome 5 introgression and therefore have Rx3 and pto but lack SLS-2, and other with recombinant introgressions so that they maintained SLS-2 and now have Rx3, but not Pto. These lineages were one generation behind the transfer of the chromosome 11 bacterial resistance genes to the Cornell blight resistant parental line, and so have only undergone one field tested for horticultural and disease resistance performance in summer 2019 (see Table 2). Denser genotyping on the CH5 materials did not indicate any residual heterozygosity not identified by low-density genotyping throughout the breeding process, indicating the usefulness of the marker-assisted backcross breeding approach used in this work to more rapidly generate near-isogenic lines. We perceived time-saving gains of ~3 backcross generations with the implementation of this tandem MAS approach, compared to MAS backcross breeding with only foreground markers about the target QTL, and traditional MAS breeding based on phenotypic selection alone.
The chromosome 5 introgression lines displayed a larger, more vigorous growth habit with internode lengths longer by approximately 20% over the recurrent parent line CU151095-146. Chromosome 5 lines also displayed later maturity (Table 2), on approximately 14-21 days later than the CH11 lines. We consider this to be significant linkage drag that will have to be broken using the recombinant materials developed during this breeding project (see methods). Table 2 shows that the chromosome 5 introgression lines from the 2019 field were lower yielding than the CH11 lines and hybrids, though the result is not statistically significant in most cases. This trend toward lower marketable yield is likely a reflection of the later maturity of the chromsome 5 lines, as fruit of these entries remain on the vine until the first frosts, and these fruit would not have been included in the marketable yield trial. These lines may also have slightly smaller fruit than the other entries in the trial (Table 2), though once again the result was not statistically different at the 95% confidence level.
Combination of Resistances
We are currently analyzing the yield trial data from the recent 2019 field season on the CH5/CH11 combined lines and so cannot yet provide accurate statistical results; these will be presented in the 2019 VBI report and in an upcoming publication on this work that will reference funding from SARE (Winter 2020). What follows are anecdotal observation on these lines from the field trials. There was little apparent difference in fruit set or size among the lines, however the later maturity of lines homozygous for the CH5 resistance region meant some fruit were still on the vine at the end of the season when the first frosts hit. Because such late-maturing fruit were not harvested, it is likely that lines with the chromosome 5 introgression will show reduced yields by our measures. Such late production may jeopardize field-based tomato production in the northern tier, where field seasons are short. However, these late season varieties may be acceptable under the extended field seasons offered by high-tunnel production systems. The F1 hybrids heterozygous for these resistances fared better, maturing approximately 7 days later than the recurrent parent. This is encouraging, as derived commercial hybrid varieties will likely be heterozygous for all four resistances. Among the 24 plants that that had brought together all of the resistances, we identified four plants which appeared superior to our eyes in terms of yield, fruit set and quality, and plant morphology. These are presently being crossed in the greenhouse to other breeding lines from the CU, NCSU, and OSU breeding programs to develop test hybrids for commercial licensing. This downstream work is funded by USDA specialty-crop grants that were made possible by the preliminary data generated in this work.
Mapping and validation of Early Blight Resistances.
Large effect EB resistance QTL, for both stem and foliar EB infection phases, were identified in our population and mapped to three chromosomes. Several additional minor effect QTL were also identified, but these were not prioritized for future work. The impacts of these QTL were confirmed in the 2018 confirmation populations. These QTL are now in the process of being fine-mapped and have been narrowed to regions of approximately 3Mbp in size. Current QTL intervals are enough for marker assisted selection as performed in this work, but refined intervals will allow plant pathologists to study this system further and elucidate the underlying resistance mechanism.
The three prioritized large-effect QTL were located on the tomato chromosomes 1, 5, and 9, respectively. The chromosome 1 QTL reduced defoliation when fixed for the CU alleles. Regression of Foliar AUDPC against the chromosome 1 QTL alleles gave a statistically significant result both years (p < .01), and reduced defoliation on average by about ~25%. The chromosome 5 QTL also impacted defoliation. When fixed for the OH allele, defoliation was reduced on average ~40% across the two years, and the QTL regression results were statistically significant (p < .01) both years. The chromosome 9 QTL mostly contributed stem lesion resistance, reducing stem lesion AUDPC by an average ~75% across both years. The effect of this QTL was also statistically significant across both years (p < 0.01). There was some indication of QTL x QTL interactions, but these were not significant at the genome-adjusted p-value threshold. Further details regarding these QTL, including a full characterization of the QTL effects and information on predictive SNPs and variant sequences will be published in an upcoming academic journal publication (Winter 2020).
Data from growers helped make breeding decisions.
As part of our grower outreach program we presented the tomato lines and fruit to grower in LI and HV. Growers were also given anonymized tomato samples to taste from the finished CH5/CH11 lines (Figure 3) that were used to help make selections. In addition, we performed two years of on-farm trials on grower fields. Grower field days offered valuable input on the traits they liked and disliked about our breeding selections.
Current and future line releases.
Table 1 below summarizes the first breeding line releases from this project.
Table 1. Performance data on the best lines bacterial spot resistant lines that are now available for licensing. Data are from a 2018 yield trial in Freeville, NY. These lines have the bacterial spot resistance gene combinations Rx-4 and QTL-11 as well as resistance to early blight, late blight (Ph2 + Ph3), Septoria (SLS-2), Verticillium (Ve) and Fusarium (I + I2). Adjusted means for horticultural traits (with Tukey letter groupings, α=0.05) and best linear unbiased predictors (BLUPs) for performance-related traits in the lines currently available for release, as well as for the previously released Cornell recurrent parent (italicized). Negative disease BLUP values indicates that a plant is more resistant; positive BLUP value indicates more susceptible.
|
|
Horticultural Performance |
Disease Severity for Three Bacterial Spot Species |
||||
|
Source Seed Lot |
Average Marketable Fruit Weight (grams) |
Relative Maturity (1 = early, 3 = late) |
Avg Marketable Yield for a 4-plant plot (kilograms) |
X. perforans (BLUP) |
X. euvesicatoria (BLUP) |
X. gardneri (BLUP) |
|
181054-121 |
157 a |
1.25 a |
21.77 a |
-0.92 |
-0.70 |
-0.65 |
|
181055-057 |
175 a |
1.75 a |
23.76 a |
-0.92 |
-0.31 |
0.36 |
|
181055-082 |
167 a |
2.00 ab |
22.70 a |
-0.69 |
-0.37 |
-0.24 |
|
181055-133 |
172 a |
1.75 a |
20.77 a |
-0.58 |
0.32 |
-0.24 |
|
181065-180* |
113 b |
3.00 b |
28.20 a |
-0.69 |
-0.20 |
-0.44 |
|
CU151095-146 |
178 c |
2.25 ab |
14.40 b |
0.35 |
0.64 |
-0.04 |
*181065-180 has an extraneous introgression on chromosome 2 that reduces fruit size.
Table 2. Preliminary performance data on lines and hybrids developed during this project. Except for the lines also in Table 1, these breeding materials have not yet been released to the breeding community. Data are from a 2019 yield trial in Freeville, NY. The CH5 lines have the bacterial speck (pto) and spot (Rx-3) resistance genes. The CH11 lines have additional bacterial spot resistances (Rx-4 and QTL-11). The hybrids are heterozygous for all four bacterial resistance genes. All lines have resistance to early blight, late blight (Ph2 + Ph3), Septoria (SLS-2), Verticillium (Ve) and Fusarium (I + I2). Adjusted means are for horticultural traits (with Tukey letter groupings, α=0.05) as well as for the previously released Cornell recurrent parent (italicized) and a commercial benchmark (italicized).
|
Source Seed Lot |
Average Marketable Fruit Weight (grams) |
Avg Marketable Yield for a 4-plant plot (kilograms) |
Maturity Slope* (lower values are earlier maturing) |
|
181244-3 (CH5 Line) |
161.89abc |
17.62a |
0.113 cd |
|
181245-1 (CH5 Line) |
171.42abcd |
22.51ab |
0.118 cd |
|
181246-3 (CH5 Line) |
154.08a |
19.99 a |
0.126d |
|
181054-121 (CH11 Line) |
166.22abcd |
28.44ab |
0.049ab |
|
181055-057 (CH11 Line) |
192.04de |
29.86ab |
0.102 bcd |
|
181055-082 (CH11 Line) |
179.21abcde |
34.24b |
0.073abcd |
|
181241 x 181244 (F1) |
158.35ab |
28.19ab |
0.102 bcd |
|
181241 x 181245 (F1) |
190.07cde |
27.53ab |
0.098 bcd |
|
181241 x 181246 (F1) |
161.99abc |
26.91ab |
0.101 bcd |
|
181242 x 181244 (F1) |
156.95a |
30.11ab |
0.111cd |
|
181242 x 181245 (F1) |
172.99abcd |
29.20ab |
0.110cd |
|
181242 x 181246 (F1) |
188.09bcde |
23.39ab |
0.105 bcd |
|
CU151095-146 |
170.42abcd |
20.56a |
0.088bcd |
|
Commercial Benchmark |
203.74e |
25.28ab |
0.099 bcd |
*Maturity slope is calculated by fitting a linear model prop_harvest ~ date + e, where "prop_harvest" is the proportion of the total harvest for an entry collected on a particular harvest date, and "date" is the dates of harvest (in order) over the course of the season.
Table 3. Farmer taste test results. The following entries were included in a farmer taste test at the Long Island Research Station in summer 2019. Below are the average scores for the entries. The favorite, in terms of fruit size and shape, was the CH5 x CH11 hybrid 181241 x 181246, while it appears that growers preferred the internal characteristics and taste of the recurrent parent control line, suggesting some linkage drag associated with the resistance introgressions. Further breeding is now being done to improve fruit flavor and texture characteristics.
| Entry | Size (1-5)* | Shape (1-5)* | External Appearance (1-5)* | Internal Appearance (1-5)* | Taste (1-5)* |
| CU151095-146 | 3.6 | 3.9 | 3.9 | 4.4 | 4.0 |
| 181242 x 181244 | 3.8 | 4.2 | 4.1 | 3.7 | 2.8 |
| 181241 x 181246 | 4.4 | 4.3 | 4.1 | 3.8 | 3.0 |
| 181055-057 | 3.7 | 3.7 | 3.9 | 3.9 | 3.2 |
| 181245 | 3.4 | 3.6 | 3.6 | 3.7 | 3.2 |
*1 is least and 5 most preferred.
Future releases planned.
Chromosome 5 lines: Development of lines that combine the Pto (bacterial speck) and Rx-3 (bacterial spot) resistances was completed fall of 2018. The lines were trialed during the summer of 2019, so that we can select lines for release by early 2020.
Combined Chromosome 5/Chromosome 11 lines: Lines that combine all the bacterial speck and spot resistance genes and QTL (on chromosomes 5 and 11) with the fungal/oomycete resistances are being developed. The first F1 hybrids with all the bacterial resistances combined in the heterozygous state were trialed during the 2019 field season, and we anticipate having homozygous lines available for release by the fall of 2020, or perhaps sooner.
- Tandem and foreground and background genome selection can be used to rapidly introgress disease resistances from unadapted sources into modern elite breeding lines. We successfully used this approach to make near-isogenic lines in just two backcrosses with relatively small genotyping costs. Costs were cut by conducing foreground and background genome selection separately and by using low density markers. Subsequent high-density genotyping confirmed the efficacy of the approach. This method can be used by breeders to more efficiently develop disease resistant fruit and vegetable varieties adapted to the Northeast US.
- Resistance to Xanthamonas sp. causing bacterial spot was demonstrated in an elite fresh market breeding line. The chromosome 11 introgressions (bearing QTL-11 and Rx-4) had a positive impact on yield, and reduced internode lengths, producing a more compact fresh market growth habit. The chromosome 5 introgressions (bearing pto and Rx-3) increased internode lengths and led to later fruit maturity, which may pose a growing challenge in our clime. However, hybrids with the resistances did not show significant linkage drag on commercially important traits. Hybrids developed from these breeding lines are being further tested and will be made available to growers. Improved genetic resistance to bacterial speck and spot in a fresh market tomato that already bears resistances to early blight, late blight, and other important diseases offers an important complement to chemical disease control. This helps to improve the sustainability of tomato production in the Northeast and to protect growers from crop losses.
- This project required that early blight resistance loci were mapped in the tomato genome. We mapped these loci and used the new molecular markers to produce tomatoes that have enhanced resistance to early blight. These markers can also be used by other breeders to produce tomato varieties with improved early blight resistance. Early blight remains a major issue for growers in the Northeast, and our work helps to address this issue.
Education & Outreach Activities and Participation Summary
Participation Summary:
Completed activities:
- 2 farm demonstrations / field day presentations (August 2018 and 2019) at the Thompson research farm in Freeville, NY showcasing the isogenic bacterial speck and spot resistant breeding lines in progress.
- 2 farm demonstration / field day presentations (August 2018 and 2019) at the Terwilliger research farm in Freeville, NY showcasing the effects of early blight resistance QTL which were identified in the course of completing this project.
- 4 field day presentations (Events in LI and HV in August 2018 and 2019) to growers and extension showcasing the new materials.
- Two industry-focused VBI reports covering work completed to date and ongoing projects, as well as information and listings on tomato breeding lines which are currently, or will shortly be available for licensing.
- Two presentations to the Tomato Breeder's Round Table (2018 and 2019) about this funded research.
- A presentation to the Cornell Plant Breeding community about this funded research (2019).
- A Jan 2019 presentation to the Empire State Agricultural Expo in Syracuse NY.
Upcoming activities:
- One additional industry-focused VBI report (Summer 2020) covering ongoing work and breeding materials which will become available for licensing + 2 additional Vegetable Breeding institute field day presentations in Summer 2020.
- Two academic journal publications concerning this work (Late 2019 / Early 2020).
- 2+ upcoming presentations (including an exit seminar) to the Cornell community which are recorded and uploaded onto the College of Agricultural Science's YouTube page (Through late 2020).
- Additional lines are to be developed and marketed to growers and seed companies to help disseminate the results of this funded research. Advertisement avenues include those listed above (Cornell Cooperative extension, the Vegetable Breeding Institute, the Empire State Agricultural Expo, and the Tomato Breeder's Round-table).
Supplemental materials related to outreach and education
Anderson TBRT Presentation 2018
Gardener Open House Handout: Cornell Disease Resistant Tomatoes
Project Outcomes
There is increasing demand for "low-spray" or "no-spray" tomatoes, and the varieties released in cooperation with the Cornell tomato breeding program help growers to deliver this product while also sustaining a business. Critical to this effort is our ongoing work to boost disease resistance in tomato. These disease resistances can make a significant impact on grower profits by reducing crop losses when growing conditions are adverse. The tomato germplasm generated from this work is an important part of our ongoing efforts to develop Northeast-adapted varieties that growers can trust. Furthermore, the molecular markers and QTL mapped in this work will help breeding programs (including our own) to bolster early blight and bacterial disease resistance in regionally-adapted tomatoes.
As a result of this work, we have begun to use (the now validated methods of) tandem foreground and background genome selection on a regular basis in our breeding program. We have also become more aware of the need to bring "heirloom" appearance and taste traits into the hybrids we develop as a result of collaboration and tasting sessions with growers. The work conducted herein has also opened new avenues of research into the mechanism(s) and genomic location(s) of early blight resistance. We now have data on the effectiveness of bacterial speck and spot resistances in our fresh market background that we are using to make better tomato breeding decisions. All of this knowledge was critical in writing successful USDA specialty-crop grants and a USDA predoctoral fellowship grant that builds upon this work. These projects are funding downstream efforts to test and release marketable hybrids bearing our new disease resistances. We are actively using the breeding lines in these projects that were developed in part through funding from Northeast SARE.
The preliminary early blight QTL mapping made possible by this work opens up many relevant research questions. This includes: 1) Where exactly are the resistances within our QTL confidence intervals?, 2) What are the effects of these resistances on maturity?, 3) Where did these resistances originate, and 4) How do these resistances work mechanistically? We hope to address some of these questions with our ongoing work.