Final report for GNE19-208
Project Information
The importance of decentralized farmer and gardener seed saving networks to crop diversity is being increasingly recognized in the face of highly centralized formal plant breeding and seed production regimes in the Global North. However, most diversity analyses examine divergence between varieties or landraces only, and seed saving of varieties on commercial farms is now the exception rather than the rule in Western countries such as the United States. As a case study, 18 sources of ‘Jacob’s Cattle’, a traditional Northeastern variety of common bean (Phaseolus vulgaris L.) that has been cultivated in North America for centuries, were obtained from decentralized farmer and seed saver networks and evaluated for intra-varietal genetic and phenotypic diversity. Using Genotyping-by-Sequencing (GBS) as well as physiological markers evaluated in a field setting, significant levels of genetic and phenotypic divergence between seed sources were observed, and high levels of correlation were observed between genotypic and phenotypic diversity for some traits. A subset of these seed sources seem to have undergone independent outcrossing events and subsequent selection, resulting in divergent varietal strains across the network. These results with the traditional variety ‘Jacob’s Cattle’ suggest that decentralized crop stewardship networks in North America, for example the Seed Exchange network managed by Seed Savers Exchange, generate and steward significant levels of crop genetic diversity over time. Seed sources from the seed saver network exhibited diversity not represented by commercial seed sources or the USDA National Plant Germplasm System (NPGS). Overall, these results can inform the work of seed savers, farmers, and plant breeders in the Northeast and beyond.
1) Identified and obtained distinct seed sources of one traditional bean variety (“Jacob’s Cattle”) sources using regional seed companies and seed saver networks. These traditional varieties have been grown in the Northeast and throughout the United States for over 250 years. Our hypothesis was that In situ cultivation of seed sources in diverse locations over many decades led to potentially significant genetic divergence, creating a situation of low intra-source variation and high inter-source variation within one culturally defined variety. This is supported by anecdotal evidence and variation in morphology.
2) Characterized genetic diversity of collected seed sources and analyze population structure. We tested the hypothesis that intra-varietal diversity of the “crowdsourced” 'Jacob's Cattle' seed sources will be greater than multiple sources of a commercial cultivar check, the light red kidney variety 'Cal Early'. Interestingly, a similar study of wheat landraces in France demonstrated just this result (6). These assembled populations of bean land races formed the basis of future population improvement and breeding work. Increased genetic variation of a population allows greater genetic gain through selection, and potentially greater stability of performance across different environments and abiotic and biotic stressors.
3) Evaluated phenotypic traits of single source plots in a replicated field trial, using an augmented complete block design with four blocks. These data complemented our genotypic data by demonstrating whether genetic diversity detected translates into meaningful phenotypic or morphological traits. A single replicate trial was also planted in Maine at a cooperator farm of Fedco Seeds, but no phenotypic data was collected at this site.
4) Conducted on-farm participatory selection and outreach with growers to select and improve diverse variety populations at collaborator site, Fedco Seeds. This trial took place at the farm of an experienced bean grower and long-time Fedco collaborator. These trials served as a “baby” site to the replicated trial at Cornell University, and was planned to be the basis for a grower field day for dry bean growers and seed producers to observe the experimental population and select phenotypes of interest. This would have served to facilitate participatory methods of population improvement and begin the development of a regionally improved strain. **Note: due to covid-related restrictions on research travel, the planned participatory selection and field day activities were not able to be held.
Edible dry beans were historically an important crop for the northeast region of the United States, and as a legume species are beneficial for low-input, diverse crop rotation systems. Growers and consumers are increasingly interested in regionally produced staple crops, including traditional varieties valued for their culinary, visual and agronomic traits as well as their history. However, dry bean production in the Northeast has declined sharply over the past decades, and most varieties grown by commercial northeastern growers were bred in other regions, with seed re-purchased each season from highly centralized western seed production regions. In general, decentralized, in-situ management of crop diversity by many farmers in many locations was once ubiquitous, and formed the foundation of crop domestication and improvement over millennia. In-situ crop diversity management in industrialized countries such as the United States is now largely carried out by backyard seed savers and small-scale farmers, connected by grassroots seed saving networks.
The purpose of this project was to determine the role of decentralized seed saver networks to the stewardship of important crop genetic resources that may not be represented by formal breeding programs or germplasm repositories, such as the National Plant Germplasm System (NPGS). The project also was intended to create a highly diverse "crowdsourced" population of 'Jacob's Cattle' bean to be redistributed to participating seed savers or anyone else interested, providing an opportunity for regional selection and "re-adaptation" of a popular traditional bean variety with deep historical ties to New England and the Northeast. In order to expand regional production of dry beans as a profitable crop, it is imperative to ensure that growers have access to varieties that meet their needs. This is attainable by selection within traditional germplasm, and by crossing to modern germplasm.
The secondary purpose was to convene growers of mixed experience levels to facilitate a passing of knowledge from experienced to new or prospective growers, especially regarding strategies for growing high quality bean seed in the Northeastern climate.
Expansion of dry bean production in the Northeast as well as increased profitability of production, is important to maintaining healthy rural communities as well as diverse, healthy agro-ecological systems. It is also important to regional food security by providing regionally available sources of plant-based protein.
Cooperators
Research
In September of 2019, unique seed sources were identified via several channels: 1) NPGS-GRIN; 2) the Seed Exchange portal hosted by Seed Savers Exchange, a not-for-profit organization based in Decorah, Iowa; 3) an internet search of farm-based seed company inventories; and 4) snowball sampling of farmers in Maine (USA), believed to be the historical region of origin of ‘Jacob’s Cattle’ in North America. Accessions were identified by variety name, which included either the phrase ‘Jacob’s Cattle’ or ‘Trout’. Seed samples were requested and received from 18 distinct sources, with the number of seeds received varying from approximately 30 to 1200.
Seed source type varied between home gardener, commercial seed company and commercial farm. As a result, source population sizes also varied widely but were not able to be confirmed by all sources included, so these data were not presented. Some sources had only been cultivating the variety for a few years, while one seed saver source had been growing and saving seed of ‘Jacob’s Cattle’ for 30 years. Source 706, which came from the NPGS-GRIN collection, was visually identified as a likely admixture of ‘Jacob’s Cattle’ and ‘Anasazi’, the latter of which is a Southwestern heirloom with similar color and pattern to ‘Jacob’s Cattle’ but of Middle American origin and thus highly genetically distinct from ‘Jacob’s Cattle’, which is of Andean origin. Two individuals of each type were included in subsequent genotyping, which confirmed visual identification of admixed seed as being of Middle American origin, likely race Durango. These two individuals were included in downstream analyses as ‘Anasazi’, rather than ‘Jacob’s Cattle’. Finally, as a check comparison, an attempt was made to acquire 18 sources of ‘Cal Early’ light red kidney, a standard commercial variety. Only five sources were identified and seed was acquired from each of these, a limitation which is consistent with highly centralized seed production for modern cultivars.
Sequence data for additional traditional dry bean genotypes was obtained from University of Minnesota to provide further context for the study (Swegarden, 2020). These data included five heirloom varieties and two market class checks. For four of the heirloom varieties, Jacob’s Cattle Gold, Lina Sisco’s Bird Egg, Tiger’s Eye, and Peregion, 20 individual plants, originating from pure lines that had been selected from single commercial seed sources, were genotyped. For the remaining varieties, Lariat pinto, Eclipse black and the heirloom Painted Pony, single individuals were genotyped. Passport data for genotypes included in the study can be found in Table 1.
Genotyping and genetic diversity analysis
In Winter of 2020, five seeds from each source were planted in Cornell mix media and grown in container culture in the Guterman Research Center on the campus of Cornell University (Ithaca, NY USA). Leaf tissue of the first trifoliate was collected 21 days after planting from four individuals per source for genotyping. For the purposes of subsequent field evaluations in Summer of 2020, all five plants from sources with fewer than 150 seeds were grown to maturity and harvested for the purpose of increasing seed for field trial plots. DNA was extracted using the Qiagen DNEasy 96 Plant Kit (Qiagen Inc., Valencia, CA, USA) according to the manufacturer’s instructions.
A 96-plex Genotyping-By-Sequencing (GBS) library (Elshire et al. 2011) was prepared and sequenced on a NovaSeq 6000 (Illumina, San Diego, CA, USA) with shared-lane paired‐end 150 bp reads at the University of Wisconsin‐Madison Biotechnology Center (Madison, WI, USA). Reads were aligned to the Phaseolus vulgaris genome (v2.1) (DOE-JGI and USDA-NIFA) with the “bwa” aligner in the GBSv2 pipeline in TASSEL 5 (Glaubitz et al., 2014). In VCFtools (Danecek et al., 2011), Single Nucleotide Polymorphisms (SNPs) that were not biallelic, had extreme mean read depths (<3, or > 30), low minor allele frequency (<0.05), or were missing in >40% of samples were removed. SNPs were lastly filtered to exclude loci with >10% heterozygosity to remove any other likely misaligned reads. As common bean is highly inbreeding typically (90-99%), high proportions of heterozygosity at the same loci across sources was deemed unlikely. Raw reads from the University of Minnesota GBS dataset were also aligned and SNPs called along with ‘Jacob’s Cattle’ and ‘Cal Early’ data using the same pipeline as above.
Total SNP counts within varieties and seed sources for ‘Jacob’s Cattle’, ‘Cal Early’, ‘Lina Sisco’s Bird Egg’, ‘Tiger’s Eye’, ‘Jacob’s Cattle’ Gold’ and ‘Peregion’ were tallied using TASSEL 5 (Bradbury et al., 2007). A Principal Components Analysis (PCA) of two SNP datasets were conducted in TASSEL5: 1) All varieties and seed sources 2) ‘Jacob’s Cattle’ individuals only. PCA results were plotted in ggplot2 (Wickham, 2016). A Discriminant Analysis of Principle Components (DAPC) was conducted using the ‘find.clusters’ function in the ‘adegenet’ package (Jombart, 2008) to obtain k-means group assignments for sub-populations. In the analysis, Bayesian Information Criterion (BIC) values were minimized as a statistical measure of goodness of fit.
Minor alleles of individual genotypes grouped by assigned cluster, including both homozygous and heterozygous calls, were then plotted by physical location across all eleven chromosomes to visualize SNP distribution and identify haplotype blocks. In this instance, “minor alleles” represent a genotype at a given locus that differs from that of the predominant genetic cluster identified within seed source genotypes. Based on these data, a separate plot was subsequently drawn that included representative genotypes of each of the five ‘Jacob’s Cattle’ genetic clusters as well as other commercial and heirloom varieties, to evaluate for evidence of areas of similarity between varieties and genetic races. Only SNP positions with at least one polymorphic genotype within the ‘Jacob’s Cattle’ dataset were included. This allowed greater resolution to analyze potential areas of recombination between ‘Jacob’s Cattle’ sources and highly divergent genotypes of Middle American origin, but does not allow accurate analysis of genetic divergence between ‘Jacob’s Cattle’ and Middle American genotypes. 1356 SNPs were included in this analysis. Data was plotted in ggplot2 (Wickham, 2016).
Phenotypic diversity measurements and analysis
In summer of 2020, seeds from all 18 ‘Jacob’s Cattle’ sources were planted in an augmented design with 2 replications and 4 blocks at the East Ithaca Research Farm on Cornell University campus (Ithaca, NY USA). Phenotypic data was not collected on ‘Cal Early’ accessions or any lines for which sequence data was obtained from the University of Minnesota. Four ‘Jacob’s Cattle’ seed sources selected as checks were planted in each of the four blocks, while the remaining seed sources were replicated in two out of four blocks. Individual plots consisted of two 2.2-m rows planted with a “Precision Garden Seeder” push seeder (EarthWay, Bristol IN USA) at 0.76 m spacing between rows and approximately 50 plants per plot. All seeds were inoculated with “Guard-Nâ” N2-fixing bacteria (Verdesian, Cary NC USA) immediately before planting.
Phenotypic data measured in each plot consisted of main stem length in centimeters, number of main stem nodes, number of pods per plant, total seed yield per plant in grams, and 100-seed weight in grams. Each measurement was conducted on four randomly selected plants per plot. Number of main stem nodes was evaluated at time of flowering, number of pods per plant was evaluated at time of harvest, and total seed yield and 100-seed weight was evaluated after drying of seeds at 35 degrees Celsius for seven days.
Individual seed sources were assigned to one of five genetic clusters via the DAPC analysis described above and phenotypic data was grouped by genetic cluster for analysis. For each phenotype measured in the field, a mixed linear model was fitted to the data using the lme4 package in R version 4.0.3. with genetic cluster included as a fixed effect and row-pair and column-pair as random effects (Bates et al., 2015; R Core Team, 2017). Field partition coordinates row-pair and column-pair were used as model terms due to high observed within-field variation, rather than replicate and block terms. A variogram was analyzed to examine the data for spatial correlation, but as no correlation was found an autocorrelation was not implemented (Zuur et al., 2009). Least-squares means for mean cluster trait values were calculated using the ‘emmeans’ function in the emmeans package (Lenth, 2020); Tukey’s Honestly Significant Difference (HSD) was performed using the ‘cld’ function in the multcomp package (Hothorn and Westfall, 2008) for pairwise comparisons between least squares means at the α=0.05 significance level. Phenotypic data was plotted in ggplot2 (Wickham, 2016).
Table 1. Passport data for all seed sources included.
|
Variety |
Source Type |
Source Name |
Location |
Center of Origin/Race |
|
|
Jacob's Cattle |
Andean/Nueva Granada |
|
|||
|
20-701 |
Commercial |
Jacob's Cattle |
Copake, NY |
|
|
|
20-702 |
Commercial |
Jacob's Cattle |
Lynden, WA |
|
|
|
20-703 |
Commercial |
Wink's Jacob's Cattle |
Nictaux, Nova Scotia |
|
|
|
20-704 |
Commercial |
Jacob's Cattle |
Quincy, WA |
|
|
|
20-706 |
NPGS-GRIN |
Jacob's Cattle |
Pullman, WA |
|
|
|
20-707 |
NPGS-GRIN |
Jacob's or Dutch Cattle |
Pullman, WA |
|
|
|
20-708 |
Seed Saver |
Jacob's Cattle Gasless |
Clinton, ME |
|
|
|
20-709 |
Seed Saver |
Jacob's Cattle |
Viroqua, WI |
|
|
|
20-710 |
Seed Saver |
Jacob's Cattle Gasless |
Arkansaw, WI |
|
|
|
20-711 |
Seed Saver |
Jacob's Cattle |
Eugene, OR |
|
|
|
20-712 |
Seed Saver |
Jacob's Cattle Amish |
Illinois |
|
|
|
20-713 |
Seed Saver |
Deep Red Trout |
Illinois |
|
|
|
20-714 |
Seed Saver |
Mammouth Trout |
Illinois |
|
|
|
20-715 |
Seed Saver |
Coach Dog |
Illinois |
|
|
|
20-717 |
Commercial |
Jacob's Cattle |
Exeter, ME |
|
|
|
20-718 |
Farm |
Jacob's Cattle |
Berwick, ME |
|
|
|
20-719 |
Commercial |
Jacob's Cattle |
Unknown |
|
|
|
Other Traditional |
|
|
|
|
|
|
Tiger's Eye |
Commercial |
Tiger's Eye |
Unknown |
Andean/Unknown |
|
|
Jacob's Cattle Gold |
Commercial |
Jacob's Cattle Gold |
Unknown |
Andean/Unknown |
|
|
Peregion |
Commercial |
Peregion |
Unknown |
Andean/Unknown |
|
|
Lina Sisco’s Bird Egg |
Commercial |
Lina Sisco’s Bird Egg |
Unknown |
Andean/Unknown |
|
|
Painted Pony |
Commercial |
Painted Pony |
Unknown |
Andean/Unknown |
|
|
Cal Early |
|
|
|
Andean/Nueva Grenada |
|
|
20-721 |
Breeder |
Cal Early |
California |
|
|
|
20-722 |
Commercial |
Cal Early |
Idaho |
|
|
|
20-723 |
Commercial |
Cal Early |
California |
|
|
|
20-724 |
Commercial |
Cal Early |
Unknown |
|
|
|
20-751 |
Commercial |
Cal Early |
Idaho |
|
|
|
Eclipse |
Commercial |
Eclipse |
Unknown |
Middle American/Mesoamerican |
|
|
Lariat |
Commercial |
Lariat |
Unknown |
Middle American/Durango |
|
Results
After filtering, a subset of 1225 SNPs across 18 sources and 69 total individuals were utilized for genetic analysis within the ‘Jacob’s Cattle’ populations only. For analyses that included ‘Cal early’ check and University of Minnesota accessions, 9093 SNPs were used.
The number of SNPs within individual ‘Jacob’s Cattle’ sources ranged from 1 to 93. In general, the number of SNPs was substantially less within sources compared to the pooled ‘Jacob’s Cattle’ data (Table 2). Across all five ‘Cal Early’ sources, 624 SNPs remained after filtering, and within-source SNP counts ranged from 1 to 111. SNP counts within the four single-source heirloom varieties were overall lower than ‘Jacob’s Cattle’ or ‘Cal Early’, with the exception of ‘Peregion’. 72 SNPs were identified within ‘Lina Sisco’s Bird Egg’, 163 within ‘Jacob’s Cattle Gold’, 62 within ‘Tiger’s Eye’ and 2119 within ‘Peregion’. Substantially higher SNP counts within ‘Peregion’ are likely due to its identification as a seed mixture with two distinct seed phenotypes (Swegarden, 2015).
Table 2. Total SNP counts within varieties and seed sources. 18 distinct seed sources of ‘Jacob’s Cattle’, 5 distinct sources of ‘Cal Early’ light red kidney, and single sources of ‘Lina Sisco’s Bird Egg’, ‘Tiger’s Eye’, and ‘Peregion’ were included in the analysis. SNPs were identified via Genotyping-by-Sequencing (GBS). Only individuals with multiple individuals genotyped are included in this table.
|
Variety |
Source ID |
Number of Individuals |
Number of Seed Sources |
Number of SNPs |
|
Jacob's Cattle |
all |
70 |
17 |
1403 |
|
20-701 |
4 |
1 |
9 |
|
|
20-702 |
4 |
1 |
8 |
|
|
20-703 |
4 |
1 |
54 |
|
|
20-704 |
4 |
1 |
3 |
|
|
20-707 |
4 |
1 |
10 |
|
|
20-708 |
4 |
1 |
63 |
|
|
20-709 |
4 |
1 |
1 |
|
|
20-710 |
4 |
1 |
14 |
|
|
20-711 |
4 |
1 |
8 |
|
|
20-712 |
4 |
1 |
93 |
|
|
20-713 |
4 |
1 |
15 |
|
|
20-714 |
4 |
1 |
35 |
|
|
20-715 |
4 |
1 |
8 |
|
|
20-717 |
4 |
1 |
19 |
|
|
20-718 |
4 |
1 |
13 |
|
|
20-719 |
4 |
1 |
8 |
|
|
‘Cal Early’ |
all |
19 |
5 |
624 |
|
20-721 |
4 |
1 |
82 |
|
|
20-722 |
4 |
1 |
111 |
|
|
20-723 |
4 |
1 |
70 |
|
|
20-724 |
3 |
1 |
1 |
|
|
20-751 |
4 |
1 |
71 |
|
|
‘Lina Sisco’s Bird Egg’ |
single source |
19 |
1 |
72 |
|
‘Jacob's Cattle Gold’ |
single source |
20 |
1 |
163 |
|
‘Tiger's Eye’ |
single source |
20 |
1 |
62 |
|
‘Peregion’ |
single source |
20 |
1 |
2119 |
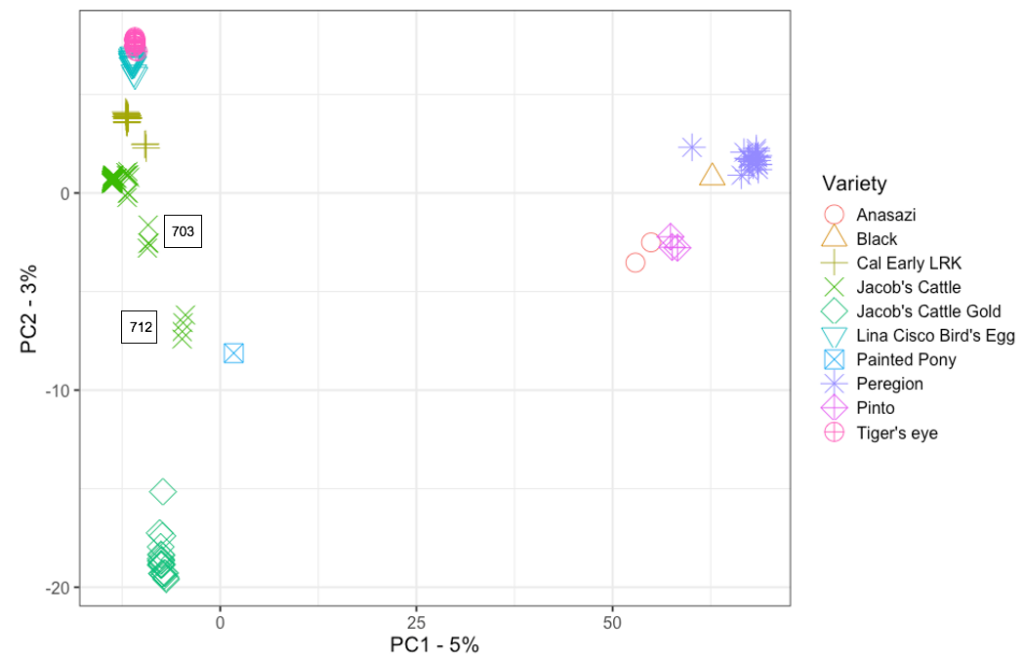
A Principal Components Analysis (PCA) of ‘Jacob’s Cattle’ with ‘Cal Early’ and other heirloom beans showed differentiation along Principal Component 1 (PC1), with two identifiable clusters. From the relative positions of known check varieties, genotypes associated with higher mean PC1 values can be assigned to the Middle American gene pool, while Andean genotypes are associated with lower PC1 values. PC1 accounted for 5% of total variance (Figure 1). PC2 primarily differentiated between sub-groups within Andean and Middle American gene pools. Within the Middle American cluster, ‘Peregion’ clusters more closely with ‘Eclipse’ than ‘Lariat’, indicating that ‘Peregion’ likely belongs to the Mesoamerican race. ‘Jacob’s Cattle’ shows greater overall differentiation between seed sources when compared to the five ‘Cal Early’ sources. Three out of four single-source heirloom varieties, ‘Tiger’s Eye’, ‘Lina Sisco’s Bird Egg’, and ‘Jacob’s Cattle Gold’, show little differentiation between individuals, indicating low diversity within single seed sources. The exception is ‘Peregion’, which is a Middle American variety that has been previously characterized as a mixture, and would therefore likely demonstrate higher diversity within source (Swegarden, 2015). The contextualization of ‘Jacob’s Cattle’ seed sources within a PCA of other traditional bean varieties and market class checks demonstrates the scale of divergence between ‘Jacob’s Cattle’ sources, with evidence of greater intra-varietal differentiation between some ‘Jacob’s Cattle’ sources compared to differentiation between distinct varieties.
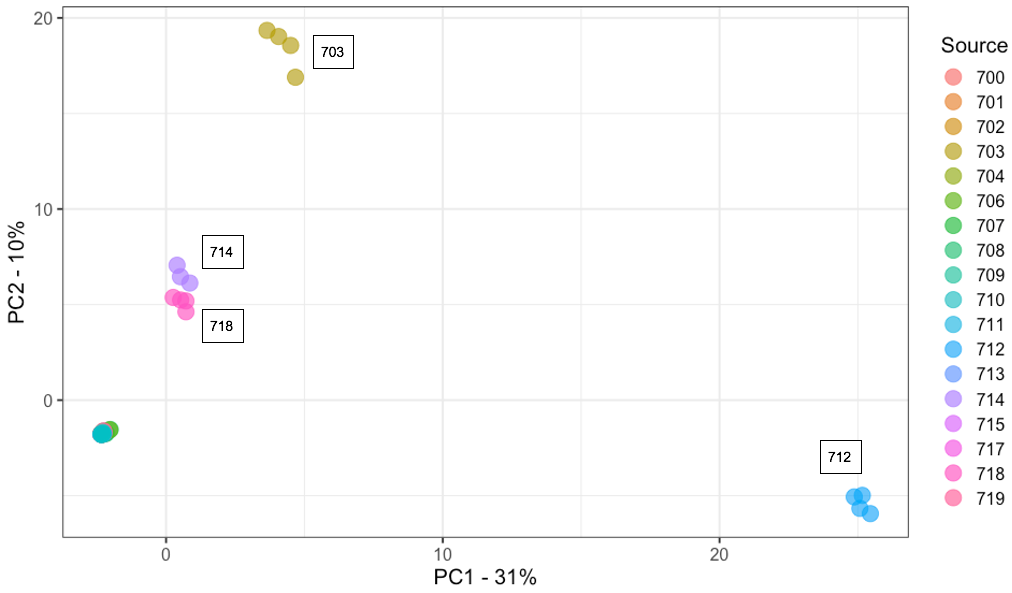
Figures 1 a-b. Scatter plot of PC1 and PC2 values for Principal Component Analysis (PCA) of a) ‘Jacob’s Cattle’ (18 sources), ‘Cal Early’ (5 sources), 4 single-source heirloom varieties, and two market class checks, black and pinto, and b) ‘Jacob’s Cattle’ seed sources only. Color of data point indicates seed source origin. PCA of 18 JC seed sources were obtained using GBS data analyzed in TASSEL5 (Glaubitz et al. 2014)
a)
A second PCA of ‘Jacob’s Cattle’ seed sources further demonstrates differentiation within ‘Jacob’s Cattle’ germplasm (Figure 1b). Fourteen seed sources tightly cluster in the lower left-hand corner of the plot, indicating little genetic differentiation. Four other sub-populations, however, show significant divergence between sources and a low level of divergence within source. A single seed source, 712, shows significant divergence across the PC1 axis which comprises 31% of total variance in the analysis.
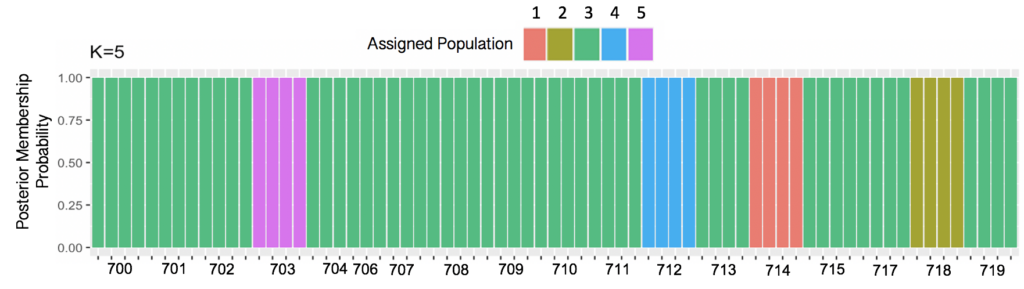
Discriminant Analysis of Principal Components (DAPC) analysis resulted k=5 genetic clusters. Groups 1, 2, 4 and 5 each consisted of a single seed source comprised of four individual genotypes. Group 3 consisted of 14 seed sources comprised of 53 individual genotypes collectively. Genetic cluster assignments were subsequently used to explore significant differences between field phenotypes and visualize SNP distributions.
Figure 2. Results from Discriminant Analysis of Principal Components (DAPC) indicate k=5 clusters across 18 seed sources using Bayesian Information Criterion (BIC) to assess goodness of fit. Results are displayed in a bar chart of posterior membership probability for each seed source across k=5 genetic clusters. 4 individual genotypes for each seed source were included, except for source 706, for which only one individual remained after filtering.
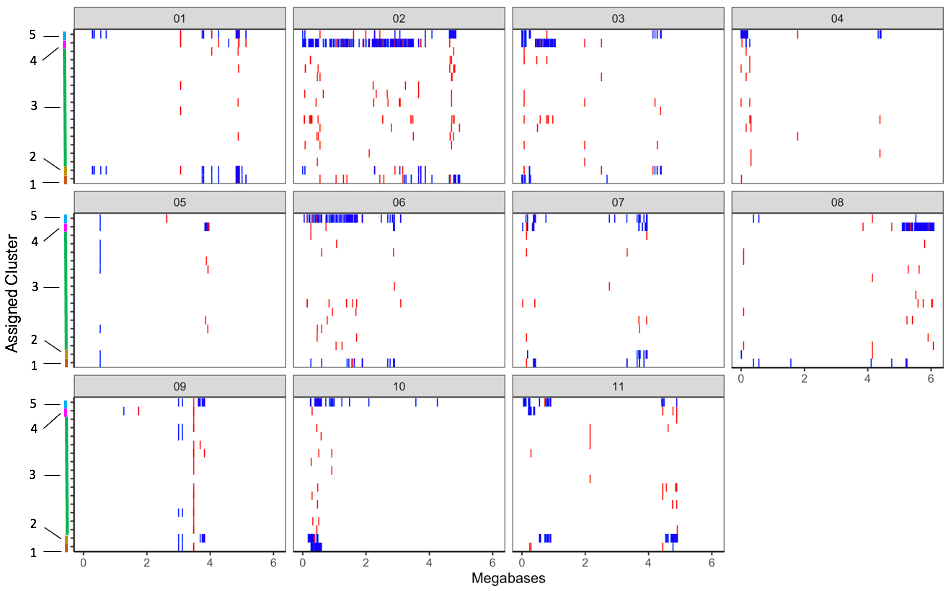
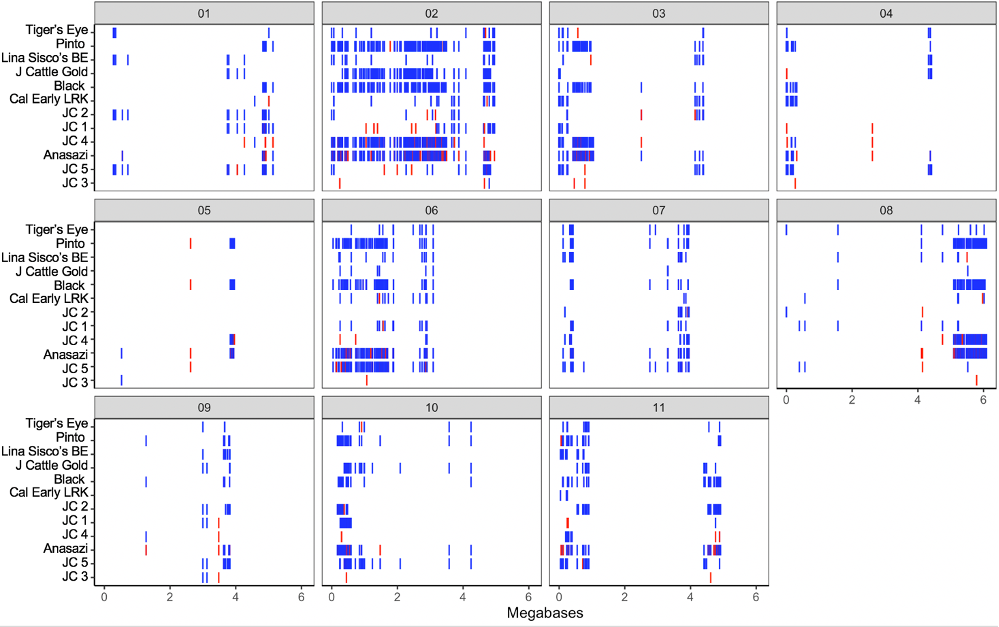
Distribution of minor alleles of individual seed sources, grouped by genetic cluster and plotted by physical location, show the presence of large minor allele haplotype blocks present in some genetic clusters, but not others (Figure 3a). In particular, clusters 4 and 5, which consist of seed sources 712 and 703 respectively, show particular haplotype divergence from the majority of seed sources represented by cluster 3. Clusters 1 and 2 also demonstrate some areas of divergent haplotypes, though blocks are less frequent and smaller in size. In the plot comparing minor allele frequency of five ‘Jacob’s Cattle’ clusters to heirloom and market class checks, minor allele haplotypes in clusters 4 and 5, which consist of sources 712 and 703 respectively, show several regions of similarity to ‘Anasazi’, black and pinto genotypes, which are of Middle American origin (Figure 3b). Minor allele distributions suggest that two ‘Jacob’s Cattle’ seed sources, cluster 4 and 5, likely outcrossed with a Middle American genotype. Cluster 4 displayed several large haplotype blocks shared with Middle American genotypes, particularly on chromosomes 2, 3 and 8. Cluster 5 displayed a large haplotype block on chromosome 6, with smaller regions homologous to Middle American checks on chromosomes 1, 2, 7, 10 and 11. These regions of shared haplotypes likely explain divergence of sources 712 and 703 seen in the PCA of ‘Jacob’s Cattle’ seed sources (Figure 1b).
Figures 3a-b. Distribution of minor allele genotype at SNPs identified across chromosomes. Homozygous calls are displayed in blue and heterozygous calls are displayed in red, for: a) all Jacob’s Cattle seed sources grouped by genetic cluster along y-axis; b) representative genotypes from each of five Jacob’s Cattle clusters, heirloom and market class checks across all chromosomes. ‘Lina Sisco’s BE’ represents ‘Lina Sisco’s Bird Egg’, ‘Cal Early LRK’ represents ‘Cal Early’ light red kidney. Exact chromosome lengths vary, for individual chromosome lengths see Phaseolus vulgaris genome v2.1 (DOE-JGI and USDA-NIFA).
a)
b)
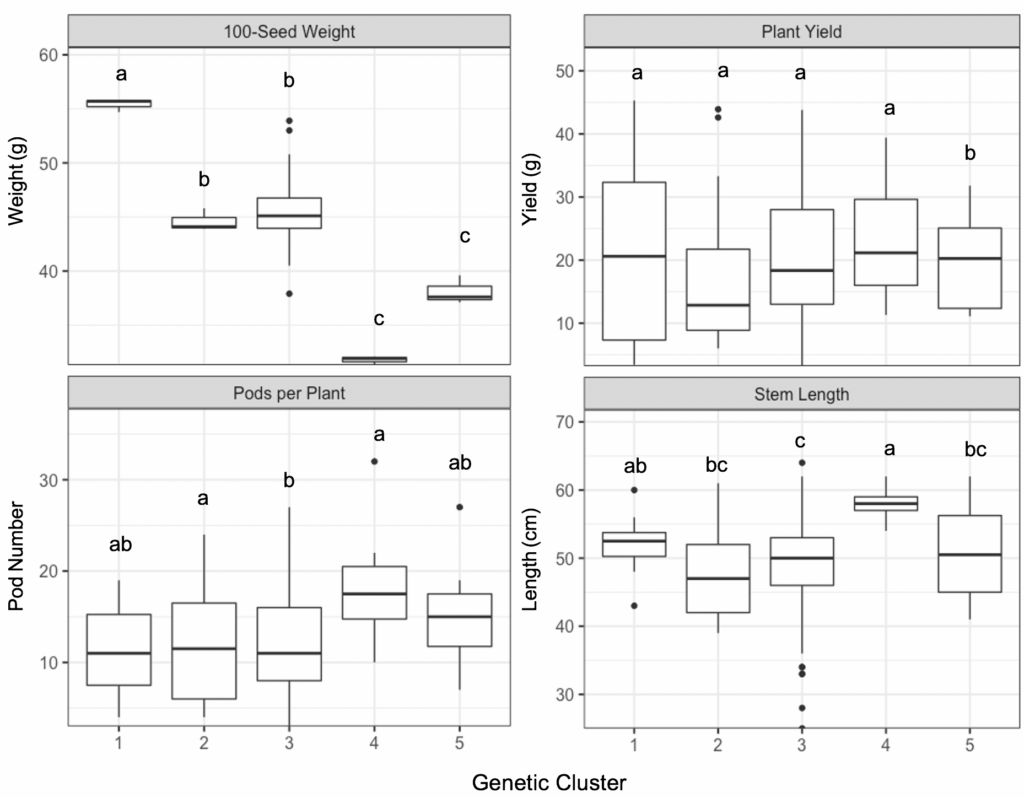
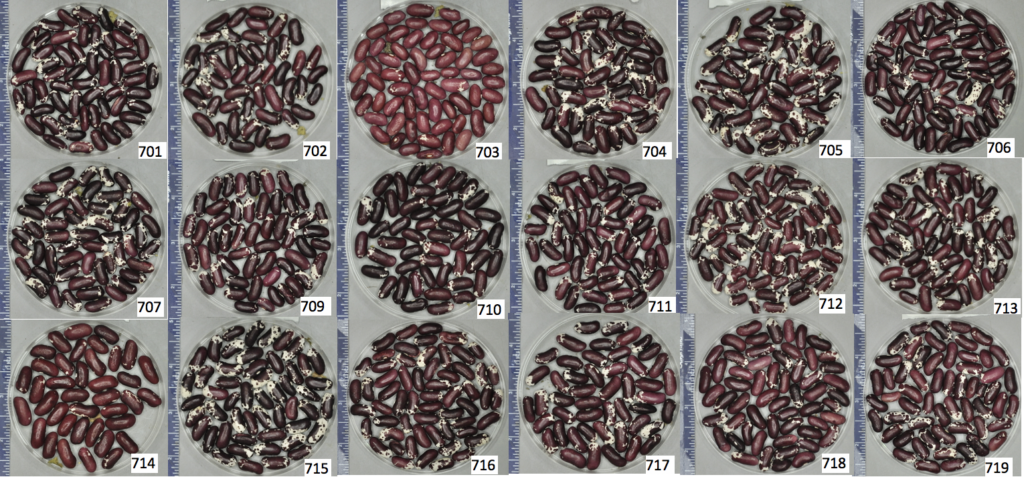
Field phenotypes demonstrated significant differences between k-means clustered group assignments for three out of five measured traits as determined using Tukey’s HSD (α=0.05). In particular, clusters 4 and 5 had significantly smaller mean 100-seed weight than the three other clusters. Cluster 4 also had significantly longer mean stem length and higher mean pod number per plant (Figure 4). No significant differences were observed for mean plant yield. Diversity in seed shape, color and pattern were also noted, though these were not measured quantitatively (Figure 5).
Figure 4. Boxplots of phenotypic trait values of 18 ‘Jacob’s Cattle’ seed sources for 100-seed weight in grams (top left), plant yield in grams (top right), pods per plant (bottom left) and stem length in centimeters (bottom right), grouped by five assigned genetic clusters. Clusters denoted with the same letter are not significantly different as determined using Tukey’s HSD (α=0.05).
Figure 5. Seed phenotypes of each of the 18 seed sources of ‘Jacob’s Cattle’ included in the experiment, labelled by source number in bottom right corner of each panel. Differences in seed phenotypes can be observed.
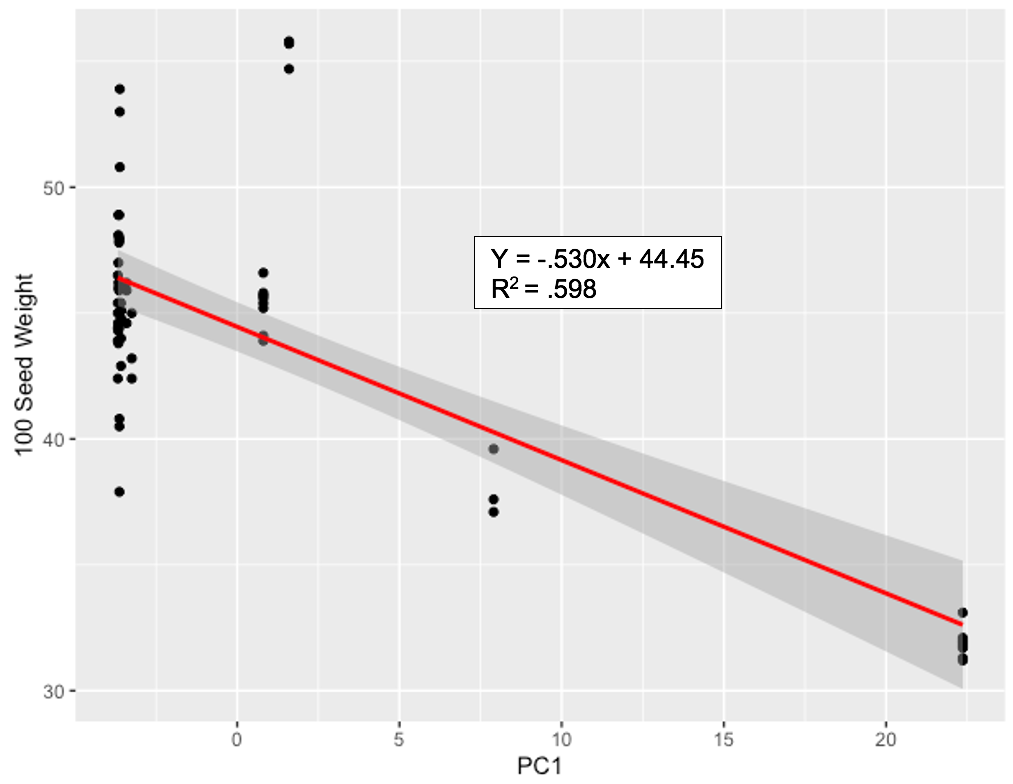
A significant relationship between mean trait value for 100-seed weight and PC1 values for all genotypes was identified through regression, resulting in an R2 value of .60 and a p-value of 1.73 x 10-13. This further supports the hypothesis that genetic divergence is a driver in observed diversity of measured phenotypic traits across ‘Jacob’s Cattle’ seed sources, and suggests that genetic divergence occurred due to historical introduction of Middle American genetic material into one or more ‘Jacob’s Cattle’ seed sources. This admixture could be the causal factor for phenotypic indicators of Middle American origin, including smaller seed size, higher pod number and greater plant height for cluster 4, which is represented by data points with highest PC1 values (Figure 6) (Singh et al., 1991).
Figure 6. PC1 values for all 18 ‘Jacob’s Cattle’ seed sources plotted against measurements for 100-seed weight phenotypic trait.
Overall, results indicate that observed genetic diversity is correlated to phenotypic diversity. Most notable were differences in 100-seed weight, pod number and stem length, and a high correlation between mean 100-seed weight and PC1 values. Along with the presence of large haplotype blocks distributed across the eleven chromosomes that appears to account for the majority of SNPs present, these findings indicate that observed genetic and phenotypic diversity within some seed sources is likely to be the result of outcrossing. Furthermore, it appears that some instances of outcrossing occurred with genotypes of Middle American races highly unrelated to ‘Jacob’s Cattle’, which is of Andean origin.
Discussion
Polymorphism within ‘Jacob’s Cattle’ was largely observed between seed sources rather than within seed sources, indicating that genetic bottlenecks within networks of seed distribution (i.e. the exchange of very small amounts of seed), are likely commonplace. The 14 seed sources assigned to cluster 3 appeared to be highly related, which may suggest a relatively recent common seed source, or simply an absence of genetic divergence after seed sources were distributed. This cluster may offer limited potential for selection or adaptation when faced with contrasting environments. However, seed sources 712 (‘Jacob’s Cattle Amish’), 714 (‘Mammoth Trout’), 703 (‘Wink’s Jacob’s Cattle’) and 718 (‘Jacob’s Cattle’) demonstrated significant differentiation from each other and all other seed sources, forming distinct genetic clusters in the DAPC model. Sources 703 and 712 in particular show evidence of one or more historical outcross events with a Middle American genotype. While these sources still largely cluster with Andean genotypes in a PCA (Figure 1), several minor allele haplotype blocks that are shared between these sources and Middle American genotypes (Figure 3b) indicates that such a cross likely occurred. This outcrossing event may have been followed by seed saver selection back towards a ‘Jacob’s Cattle’-like phenotype that could explain the relatively low frequency of Middle American haplotypes in the current genotypes, as segregating material with Middle American traits were removed from the gene pool.
It should be noted that three out of four of the divergent seed sources were named distinctly, indicating an understanding that they represent a unique strain of the variety. However, other uniquely named strains ‘Coach Dog’, ‘Dutch Cattle’, ‘Deep Red Trout’, and ‘Jacob’s Cattle Gasless’ clustered with the majority of other seed sources. This indicates that naming of a variety or strain may or not capture meaningful strain diversity at the genetic level. Two of the four divergent sources were obtained from the Seed Saver’s Exchange network, the third from a regional farm-based seed company, and the fourth from a commercial farm in Maine. The two accessions procured from the NPGS-GRIN collection clustered with the majority of other sources in cluster 3. This finding that ex-situ germplasm conservation resources do not capture the extent of intra-varietal diversity maintained within In-situ seed saver networks is supported by similar comparisons in European seed systems (Negri and Tiranti, 2010; Enjalbert et al., 2011). ‘Jacob’s Cattle’ sources also demonstrated significantly higher diversity between sources than did the commercial kidney variety, indicating that more centralized seed production systems typical for elite cultivars may not generate as much diversity with varieties, perhaps due to stricter controls on varietal purity and identity preservation (AOSCA, 2021).
More broadly, this case study indicates that, for traditional varieties being cultivated within decentralized seed saving networks, meaningful intra-varietal diversity is likely to be found at both genetic and phenotypic levels, even in autogamous crops such as common bean. However, local adaptation via allele and haplotype frequency change is likely contingent upon either an initial population with sufficient genetic variance, or the occurrence of an outcrossing event to generate such variance. Interestingly, past findings indicate that outcrossing in autogamous crops such as common bean increases when presented with a stressful environment, perhaps one evolutionary mechanism to initiate short-term crop adaptation (Klaedtke et al., 2017). Though beyond the scope of this study, it is important to note that phenotypic divergence within crop varieties in response to environmental conditions can also occur at the heritable epigenetic level, changes that in some cases contribute to local adaptation (Galloway, 2005).
Within the context of institutional plant breeding efforts in the Global North, germplasm maintained within decentralized stewardship networks comprised of home gardeners, farmers and freelance plant breeders is rarely recognized as a significant source of genetic diversity or adaptive traits, as for instance, compared to formal germplasm repositories (Almekinders 2000; Galluzzi et al. 2010). In part, strict commodity market class standards such as canning quality may play a role in disincentivizing use of novel germplasm in formal breeding programs (Kelly, 2010). In contrast, freelance plant breeders operate primarily in organic or low-input farming systems and emphasize distinct breeding goals such as adaptation to local environment and novel flavor and culinary quality (Deppe, 2021). Freelance plant breeders have made extensive use of seed saver networks such as Seed Saver’s Exchange as well as regional farm-based seed companies in sourcing germplasm (Deppe, 2021).
Significant diversity across ‘Jacob’s Cattle’ seed sources seems to have been generated by the introduction of new genetic material into stewarded populations via outcrossing, followed by subsequent selection by one or more seed savers over the course of population stewardship. Four out of eighteen seed sources exhibited significant genetic divergence, underscoring the important role that decentralized seed saving networks play in stewarding meaningful genetic diversity within traditional crop varieties.
These findings are meaningful for seed savers, farmers and plant breeders in the Northeast that are interested in accessing diverse genetic resources of traditional crop varieties. Aggregation of multiple varietal seed sources or strains from across seed saver networks may be a useful tool to facilitate regional selection and adaptation, especially given that individual seed sources show evidence of significant genetic bottlenecks and low within-source diversity. This strategy could constitute “crowdsourcing” of intra-varietal diversity, alternately conceived of as a single varietal core collection. This strategy could be especially meaningful in cultural or geographical centers of origin, or when seeking to adapt a variety to a new environment. These results are also significant for formal breeding programs interested in using germplasm that has been managed in-situ for genetic improvement. Plant breeders may wish to take into account potential seed source divergence when evaluating germplasm in variety trials or for parental selection. Sources could either be evaluated by trialing side-by-side, or simply pooled in the hopes of increasing a baseline level of genetic diversity for future selection.
In-situ management of crop diversity within seed saver networks represents an understudied venue, and further study is needed to improve our understanding of how these complex and diffuse seed systems may influence crop adaptation to diverse environmental conditions. However, current evidence suggests that decentralized farmer and gardener seed saver networks may play an important role in protecting diversity and security of crop genetic resources, a key component of global food security.
References
Aguilar, J., Gramig, G. G., Hendrickson, J. R., Archer, D. W., Forcella, F., and Liebig, M. A. 2015. Crop Species Diversity Changes in the United States: 1978–2012. PLOS ONE 10(8):e0136580.
Almekinders, C. 2000. The Importance Of Informal Seed Sector And Its Relation With The Legislative Framework. GTZ-Eschborn.
AOSCA. 2021. Seed Certification - Association of Official Seed Certifying Agencies. https://www.aosca.org/programs-and-services/seed-certification/.
Bates, D., Maechler, M., Bolker, B., and Walker, S. 2015. Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software 67(1):1–48.
Bellucci, E., Bitocchi, E., Rau, D., Nanni, L., Ferradini, N., Giardini, A., Rodriguez, M., Attene, G., and Papa, R. 2013. Population structure of barley landrace populations and gene-flow with modern varieties. PLoS ONE 8(12).
Bhandari, B. and Gauchan, D. 2018. Intra-varietal diversity in landrace and modern variety of rice and buckwheat. Journal of Agriculture and Environment.
Bradbury, P., Zhang, Z., Kroon, D., TM, C., Y, R., and ES, B. 2007. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633-2635.
Brouwer, B., Winkler, L., Atterberry, K., Jones, S., and Miles, C. 2016. Exploring the role of local heirloom germplasm in expanding western Washington dry bean production. Agroecology and Sustainable Food Systems 40(4):319–332.
Ceccarelli, S. 1989. Wide adaptation: How wide? Euphytica 40(3):197–205.
Cichy, K. A., Porch, T. G., Beaver, J. S., Cregan, P., Fourie, D., Glahn, R. P., Grusak, M. A., Kamfwa, K., Katuuramu, D. N., McClean, P., Mndolwa, E., Nchimbi-Msolla, S., Pastor-Corrales, M. A., and Miklas, P. N. 2015. A Phaseolus vulgaris diversity panel for andean bean improvement. Crop Science 55(5):2149–2160.
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., Handsaker, R., Lunter, G., Marth, G., Sherry, S. T., McVean, G., and Durbin, R. 2011. The Variant Call Format and VCFtools. Bioinformatics 27(15): 2156–2158.
Darby, H. and Cummings, E. 2018. 2017 Heirloom Dry Bean Variety Trial.
Deppe, C. S. 2021. Freelance Plant Breeding. Plant Breeding Reviews 44(1): 113–186.
Döring, T. F., Knapp, S., Kovacs, G., Murphy, K., Wolfe, M. S., Döring, T. F., Knapp, S., Kovacs, G., Murphy, K., and Wolfe, M. S. 2011. Evolutionary Plant Breeding in Cereals—Into a New Era. Sustainability 3(10):1944–1971.
Enjalbert, J., Dawson, J. C., Paillard, S., Rhoné, B., Rousselle, Y., Thomas, M., and Goldringer, I. 2011. Dynamic management of crop diversity: From an experimental approach to on-farm conservation. Comptes Rendus - Biologies 334(5–6):458–468.
Galloway, L. F. 2005, April 1. Maternal effects provide phenotypic adaptation to local environmental conditions. New Phytologist 166(1): 93-100.
Galluzzi, G., Eyzaguirre, P., Negri, V., Galluzzi, G., and Eyzaguirre, P. 2010. Home gardens: neglected hotspots of agro-biodiversity and cultural diversity. Biodiversity and Conservation 19:3635–3654.
Gioia, T., Logozzo, G., Marzario, S., Zeuli, P. S., and Gepts, P. 2019. Evolution of SSR diversity from wild types to U.S. Advanced cultivars in the Andean and Mesoamerican domestications of common bean (Phaseolus vulgaris). PLoS ONE 14(1).
Glaubitz, J., Casstevens, T., Lu, F., Harriman, J., Elshire, R., Sun, Q., and Buckler, E. 2014. TASSEL-GBS: A High Capacity Genotyping by Sequencing Analysis Pipeline. PLoS ONE 9(2): e90346
Howard, P. 2015. Intellectual Property and Consolidation in the Seed Industry. Crop Science 55:1–7.
Hufford, M. B., Berny Mier Y Teran, J. C., and Gepts, P. 2019, April 1. Crop Biodiversity: An Unfinished Magnum Opus of Nature. Annual Review of Plant Biology (70): 727:751.
Jombart, T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24 (11):1403–1405.
Kelly, J. D. 2010. The Story of Bean Breeding: dry bean production and breeding research in the U.S. Michigan State University.
http://www.hrt.msu.edu/pbgp/Links/plantbreedingintro.html
Klaedtke, S., Caproni, L., Klauck, J., de la Grandville, P., Dutartre, M., Stassart, P., Chable, V., Negri, V., Raggi, L., Klaedtke, S. M., Caproni, L., Klauck, J., De la Grandville, P., Dutartre, M., Stassart, P. M., Chable, V., Negri, V., and Raggi, L. 2017. Short-Term Local Adaptation of Historical Common Bean (Phaseolus vulgaris L.) Varieties and Implications for In Situ Management of Bean Diversity. International Journal of Molecular Sciences 18(3):493.
Negri, V. and Tiranti, B. 2010. Effectiveness of in situ and ex situ conservation of crop diversity. What a Phaseolus vulgaris L. landrace case study can tell us. Genetica 138(9):985–998.
Phaseolus vulgaris v2.1, DOE-JGI and USDA-NIFA, http://phytozome.jgi.doe.gov/
R Core Team. 2017. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna.
Seed Savers Exchange Heirloom Seeds. 2021. https://www.seedsavers.org/.
Singh, S. P., Gepts, P., and Debouck, D. G. 1991. Races of common bean (Phaseolus vulgaris, Fabaceae). Economic Botany 45(3):379–396.
Stone, S., Boyhan, G., and McGregor, C. 2019. Inter- and Intracultivar Variation of Heirloom and Open-pollinated Watermelon Cultivars. HortScience 54(2):212–220.
Swegarden, H.R. 2020. GBS of Heirloom Common Bean Pure Lines. NCBI – SRA Bioproject ID: PRJNA667092. https://www.ncbi.nlm.nih.gov/bioproject/
Swegarden, H. R. 2015. Selection of commercial and heirloom common bean (Phaseolus vulgaris L.) for organic production in Minnesota. University of Minnesota.
Swegarden, H. R., Sheaffer, C. C., and Michaels, T. E. 2016. Yield Stability of Heirloom Dry Bean (Phaseolus vulgaris L.) Cultivars in Midwest Organic Production. HORTSCIENCE 51(1): 8-14.
Thomas, M., Dawson, J. C., Goldringer, I., and Bonneuil, C. 2011, March 1. Seed exchanges, a key to analyze crop diversity dynamics in farmer-led on-farm conservation. Genetic Resources and Crop Evolution 58(3): 321-338.
Thomas, M., Demeulenaere, E., Dawson, J. C., Khan, A. R., Galic, N., Jouanne-Pin, S., Remoue, C., Bonneuil, C., and Goldringer, I. 2012. On-farm dynamic management of genetic diversity: the impact of seed diffusions and seed saving practices on a population-variety of bread wheat. Evolutionary applications 5(8):779–95.
Veteto, J. R. 2008. The history and survival of traditional heirloom vegetable varieties in the southern Appalachian Mountains of western North Carolina. Agriculture and Human Values 25(1):121–134.
Wickham, H. 2016. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag.
Wilker, J., Navabi, A., Rajcan, I., Marsolais, F., Hill, B., Torkamaneh, D., and Pauls, K. P. 2019. Agronomic Performance and Nitrogen Fixation of Heirloom and Conventional Dry Bean Varieties Under Low-Nitrogen Field Conditions. Frontiers in Plant Science 10.
Zuur, A. F., Ieno, E. N., Walker, N. J., Saveliev, A. A., and Smith, G. M. 2009. Violation of Independence – Part II. Mixed Effects Models and Extensions in Ecology with R p. 161–191.
Education & Outreach Activities and Participation Summary
Participation Summary:
A short summary of 'Jacob's Cattle' diversity study results were presented at the Northeast Organic Seed Conference, which occurred online, in January of 2021. Approximately 120 attendees were present for the session entitled "A Resilient Seed Community" in which several stories related to the Jacob's Cattle bean were shared.
A separate session on seed production also included a presentation of organic disease management for dry bean seed crops, with approximately 120 attendees.
Sessions were also recorded and will be posted on Youtube, allowing future dissemination of information as well.
The project was also presented to a diverse audience of consumers, plant breeders, farmers and service providers at the 2020 Culinary Breeding Network Variety Showcase in Portland, Oregon. The project was paired with a chef, who prepared a culinary item using Jacob's Cattle bean. An explanation of seed saver networks and improvement of dry bean varieties for the Northeast were shared via a poster, seed demonstration and verbally. Approximately 700 people attended the event.
Finally, project results were shared as part of a department seminar on April 15th, with approximately 30 attendees comprised mostly of researchers. A recording of this seminar is available here: https://www.youtube.com/watch?v=BmiL3OC5VJw&list=PLHPXm2Es8aQAIzSUe2l_sNQQdj8chGLrW&index=17&t=2773s
A manuscript of project results has also been submitted to a peer-reviewed article for review.
Project Outcomes
I hope that this research contributes to increased recognition of the importance of informal germplasm exchange networks and the importance of this in-situ conservation for continued improvement of our crop varieties, and inspire further research in this area. I also think it can inspire farmers and home gardeners to save seeds and contribute to the long term resilience of our genetic resources. By creating a pooled population of Jacob's Cattle bean, this population can be used to re-select the variety for diverse environments, and also inspire plant breeders or seed savers to create similarly "crowdsourced" populations of diverse varietal strains or sources. This increase of intra-varietal diversity at a field or regional scale can help a variety be more yield stable, less susceptible to environmental stress and more profitable for a commercial grower. It also highlights the important fact that heirloom or traditional varieties are always evolving and are never static,
With respect to the educational products focused on bean seed production and disease management, this work can help seed growers and farmers safely and productively grow bean seed in the Northeast, which can be a profitable enterprise, as well as being an important component of on-farm breeding and selection work. Organic management can help to reduce the use of fungicides or copper-based bactericides.
This project provided me with an opportunity to conceive of and carry out a research project incorporating genomics, field research, participatory research and farmer outreach from start to finish, which was an enormous learning experience and has helped to inform and work towards my goals for a future career in research and education. I was able to also make new professional connections, including with Seed Saver's Exchange and regional seed companies here in the Northeast and in other parts of the country.
This project also required me to conduct a genomic analysis of diversity including all steps from tissue sampling to sequence filtering and data analysis, which was a steep learning curve for me but was a very useful and important experience to my plant genetics training. I think this project also influenced the thinking of my advisor and fellow lab members about how to evaluate and access germplasm, especially of heirloom varieties we might use in the breeding program, as most of us expected to find little to no diversity between seed sources.
Although I already had an interest in decentralized and farmer selection of crops for agro-ecological and regional food systems, this project allowed me to deepen my understanding of in-situ genetic resource management as well as highlight the importance of these regional and farmer-led efforts. After this experience, I am even more interested in working to help strengthen regional seed systems here in the Northeast, as it's clear how important these systems are to long term genetic diversity and cropping system resilience.
It was unfortunate that pandemic restrictions prevented us from engaging with farmers and seed savers at a field day, and conducting participatory selection at that time. Additionally, because our harvested seed from 2020 trials tested positive for quarantined pathogens, we are not able to send the bulked population back to seed savers that contributed seed, as we planned to do. I think that future similar work should include better plans to distribute the bulked population in a way that can guarantee pathogen-free seed to distribute, for example bulking larger amounts in a greenhouse, or incorporating a seed increase in a more arid environment. This would allow more integration of farmers and gardeners in the project, and increase the likelihood that the bulked population is actually adopted and grown by farmers and gardeners rather than languishing in seed storage.
I think this concept could also be extended to more diverse bulked populations. Whether generated by crosses at an institution, or as simply a bulked population of many varieties or other germplasm sources, distribution of bulked populations to farmer and gardener networks could allow relatively low-cost participatory and decentralized selection, especially in a pulse crop such as dry bean where the harvested grain is the seed, and harvest is relatively easy for growers of many scales.