Final report for GNE20-237
Project Information
Seedling diseases, like Pythium, are among the top diseases leading to lost soybean bushels each year in the Northern United States. Farmers find variability in Pythium disease outcomes in fields with similar environmental conditions and see fungicide efficacy vary without a clear cause. Since Pythium is a soilborne pathogen known to synergize with other root rot pathogens and be susceptible to some common biocontrols like Trichoderma, this study hypothesizes that this variability seen by farmers can be in part explained by differences in the soil microbiome composition, with specific key fungal and bacterial taxa driving Pythium disease outcomes during early soybean development.
To understand the soil microbiome's role in influencing Pythium disease outcomes, this study tracked shifts in soil fungal and bacterial diversity and abundance over 14 days from planting in Pythium sylvaticum inoculated soil and assessed soybean root and shoot disease severity on day 14. Non-destructive daily sampling for individual plants over this timeframe was enabled by custom 3D printed rhizoboxes. Experiments were conducted in a greenhouse, growth room, and field between May 2021-22. Environmental conditions assessed included soil texture, pH, moisture, and use of a fungicide seed treatment.
Analysis of results from MiSeq sequencing of soil DNA was used to relate soil microbiome composition to Pythium disease severity. Overall, soils in this experiment were not found to be suppressive of Pythium disease. In this short timeframe, drastic changes in the soil microbiome compositions were not seen so the initial microbiome is useful for making management decisions. Abundant bacterial phyla, such as Acidobacteriota and Verrucomicrobiota, correlating with the high disease ratings seen in these experiments can be studied further to understand their role in supporting Pythium disease severity. Their abundance in the microbiome at planting can also be predictive of Pythium disease severity that will require farmers to actively manage the pathogen, such as by fungicide application.
Overall, this study provides an understanding of soil microorganisms supporting Pythium disease in soybeans that can be alternative control targets to indirectly manage this soilborne pathogen. In time, lowering the abundance of these organisms in the soil microbiome could lead to the establishment of a Pythium suppressive soil which would provide long-term sustainable disease management. This would minimize the need for toxic chemical controls that reduce farmer profit and drive pathogen resistance.
Furthermore, this project provides an educational resource on the methods and challenges involved in agricultural microbiome research for future scientists. In particular, these experiments showcase the value of 3D printing as a tool to support specialized project needs and research reproducibility. This encourages the adoption of novel tools in agricultural research for conducting unique projects that support farmers in sustainable crop production.
Objective 1: Determine how environmental conditions relevant to PA soybean farmers, including soil texture, pH, moisture, and fungicide use, impact shifts in soil bacterial and fungal communities during early soybean development under Pythium disease pressure using greenhouse and field experiments.
Objective 2: Determine how shifts in soil microbiome fungal and bacterial diversity and abundance during early soybean development relate to Pythium disease severity 14 days after planting.
Objective 3: Identify key fungal and bacterial taxa in the soil microbiome driving Pythium disease outcomes under the different environmental conditions that can be studied further to develop new sustainable alternative Pythium management strategies.
Pythium is a soilborne seedling pathogen causing germination failure and root rot in multiple economically important crops like corn and soybean, which are found in rotation in the Northeastern US. It is especially problematic in the Northern US due to the wet weather and cooler conditions at planting that promote disease development. Pythium is managed using fungicide seed coatings; however, these add to farmer input costs and do not always perform well. Identifying reliable alternative management strategies for integrated pest management of seedling diseases like Pythium lessens the need for fungicide applications that lower farmer profits, have negative health consequences, and can lead to pathogen resistance.
The purpose of this project is to develop a foundational understanding of the soil microbiome during the early soybean development stage under Pythium disease pressure to identify key microorganisms driving disease outcomes in individual plants that provide alternative targets for sustainable Pythium disease management. It considers several soil conditions seen in Northeastern US agriculture so that results are relevant across this region. Furthermore, this project seeks to understand unintended fungicide effects on the soil microbiome that may be impacting key species driving Pythium disease outcomes, which can provide farmers an additional reason not to apply fungicides. Future research can build off of this project's methods and results to develop biocontrols and other alternative sustainable management strategies that synergize with the natural microbiome to minimize Pythium disease. Overall, supporting a disease suppressive soil microbiome is an environmentally conservative way to manage soilborne seedling diseases like Pythium and promote sustainable agricultural production in the Northern US for the long term.
Research
Experiment Design Overview
See subsequent sections for more details on treatments, rhizoboxes, soil preparation, inoculation method, disease severity rating, sample collection, sequencing & analysis.
Experiments were conducted at the Penn State Greenhouse Complex on the University Park Campus and Rock Springs Research Farm. Table 1 below provides details on the differences across experiments. All rhizoboxes only had one (1) seed in them and the round pots had six. Rhizoboxes were numbered and color tagged by treatment to ensure they were set up correctly with the proper soil and treatment amendments. Pots were included in the first experiment to provide additional post-emergence disease data in case significant germination failure or non-emergence occurred. However, this ended up not being an issue and the pots added to the intensity of the experiment for the small research team so these were not included in the subsequent growth room experiment. These pots do provide an additional point of comparison for field experiment row plants, which were also grown in close proximity to other plants, that underwent disease rating.
In the field experiment, the (3) plot sites were measured to the same size in a flat area in the middle of a larger field selected based on the soil pH. There were two experimental rows per treatment, with a buffer row of soybeans planted between them and on the outer sides. Holes were dug in the plot site at equidistant intervals within the rows and soybeans were planted in the rows between the boxes. The soil from each hole filled the specific rhizobox that was placed there in the sheath that was implanted in the hole. Excess soil filled the gaps between the sheath and the hole edges for a snug fit.
The greenhouse, which had windows opening to the outdoors, and field experiments had less environmental control over temperature and humidity. The growth room was fully indoors and allowed for environmental conditions more favorable to Pythium disease. For the indoor experiments, plants were watered as needed based on visual assessment and soil moisture readings, but only soil moisture treatment plants received daily watering. All plants were watered at the same time. Soil moisture was measured but only soil moisture treatments received water (daily) in the field experiment, as soybean farmers would normally rely on nature and not irrigate fields. Two Bluetooth Govee Dual Temperature & Hygrometer Readers were set on opposite sides of the table in the indoor experiment and data was downloaded after harvest on Day 14. Button sensors were set in the middle of each (3) field plot, as the Govee Devices are for indoor use. These devices allowed for a continuous record of temperature and humidity over the experiment timeframe. Plants were randomly assigned to a new location in the greenhouse each day to avoid location bias, although all plants fit on a single table in the indoor experiments and were in close proximity to each other regardless. In the field, plants were returned to their specific numbered sheath implanted into the soil.
Plants were grown for 14 days from planting and harvested on day 14. After final soil samples were collected and plants were harvested, leftover soil was discarded and the rhizoboxes cleaned and bleach sterilized in preparation for the subsequent experiment.
Table 1. Experiment Set-Up Overview
| Experiment | Timing | # Rhizobox Treatments & Controls | Other Plants | Notes |
| Greenhouse | May 2021 |
(3) sterile, uninoculated soil per each (4) soils (3) sterile, inoculated soil per each (4) soils (4) non-sterile, inoculated per each (7) treatments (2) extra silty soil treatment (proxy control), non-sterile, inoculated Rhizoboxes= 54 |
(1) pot, with 6 seeds 2" apart per (15) treatment/control sets In-Pot plants=90 |
Disease ratings of plants in & final soil sample collection from pots occurred on Day 15, due to time constraints on Day 14. |
| Growth Room | April 2022 |
(3) sterile, uninoculated soil per each (4) soils (3) sterile, inoculated soil per each (4) soils (4) non-sterile, inoculated per each (7) treatments (2) extra silty soil treatment (proxy control), non-sterile, inoculated Rhizoboxes= 54 |
None | This room was fully indoors, allowing better environmental control than the greenhouse. |
| Field | May 2022 |
(4) non-sterile, uninoculated per each (3) treatments (4) non-sterile, inoculated per each (6) treatments (2) extra silty soil treatment (proxy control), non-sterile, inoculated Rhizoboxes= 38 |
(4) samples collected directly from field row of soybeans between rhizoboxes per (6) treatment/ (3) control (2) extra field sample plants from silty soil treatment (proxy control) In-Field Row Plants=38 |
There were 3 different field sites representing the (2) different pH levels and (1) proxy control soil.
|

Treatments
This experiment considers four different management or soil conditions present in PA soybean production systems: soil type, pH level, soil moisture, and fungicide seed treatment. These were selected based on environmental factors that support Pythium disease development and are described in Table 2 below.
Table 2. Treatment Overview
| Treatment | Description | Notes on Why Included |
|
Soil Texture
|
Silty clay soil versus a sandy loam soil. Silty clay soil was the base soil used for treatment comparison (proxy control for inoculated boxes) & used in boxes for Fungicide and Moisture Treatments. |
Silty soils are better at retaining moisture and have higher Pythium incidence than silty soils. Silty soils are common in PA. |
|
Fungicide
|
Allegiance seed treatment applied, as directed by label. This targets oomycete pathogens like Pythium. |
Ranges in fungicide efficacy are seen by farmers and this study seeks to see if this can be explained by the specific microbial communities present. This study also explores how fungicides impact non-target organisms that may play a role in health outcomes or plant development. |
| Soil Moisture |
Daily watering to drip through excess; soil moisture probe reading & visual assessment confirmed moisture level was different than other treatments. Other plants were watered as needed based on visual assessment and probe reading, but not daily. |
Production fields in PA are not always flat and this can allow water to collect in portions of the field, stressing plants and providing a conducive environment for Pythium disease. Climate change could also increase precipitation seen in PA. Pythium is an oomycete, or water mold, and water plays an important role in its ability to germinate, spread and cause disease. |
| Soil Moisture & Fungicide | Allegiance seed treatment applied, as directed by label, & daily watering. Soil moisture probe reading & visual assessment confirmed moisture level different than other treatments. | Soil moisture plays a role in the fate of fungicides and their efficacy. |
| Soil pH | High and low pH soil collected based on historical tests and verified by soil fertility testing. Actualized indoor soil pH was 7.3, 7, and 6.7 (proxy control). Actualized field pH was 5.2, 6.15, and 6.22 (proxy control). |
Soybeans and Pythium both prefer a pH range of 6-7. This allows us to have multiple soils with varied starting microbiomes to see general trends across microbiomes. |
|
Sterile Controls (Indoor only) |
Each soil (2 pH & 2 soil texture soils) was autoclaved separately and used for these controls. Half of sterile boxes per soil were inoculated with Pythium and the others were uninoculated. |
Considers plant development with limited microbiome effects for baseline plant growth in a disease-free environment versus growth under Pythium disease pressure. Note that as a legume with a symbiotic Rhizobia relationship, soybean development can be hindered by this sterilization. |
Rhizoboxes
The novel use of 3D printed rhizoboxes has the advantage of enabling multi-day non-destructive rhizosphere soil sample collection. Boxes can be use over multiple experimental runs with bleach sterilization, as the filament would melt in an autoclave. The filament is biodegradable polylactic acid (PLA), which is also more environmentally friendly to produce than other plastics. Boxes were designed using Tinkercad and 3D printed at the Media Commons at the Penn State University Library. 3D printing the boxes allowed for customization and uniformity of the final boxes. The design file can also easily be shared with other researchers for reproducibility.
Video of rhizobox being printed at the Penn State Libraries Media Commons, from 3DPrinterOS
Video of rhizobox sheath being printed at the Penn State Libraries Media Commons, from 3DPrinterOS
These opaque 2 (l) x 2 (w) x 5.5 (h) inch square boxes were designed based on the specific project needs, such as number of sampling holes, and expected soybean growth over the experiment timeframe for dimensions. A grid of small holes at the bottom allows for water drainage. Pieces of bleach-sterilized plastic straws cut in half and 'ribbed' (slightly cut in the middle so water or microbes could pass though) were placed in holes in the side of the box, protruding slightly on each side so they could be easily pulled out. Straw size selected was based on the soil sample size to have enough for DNA extraction (500 mg). Care was taken when adding soil to the boxes to ensure the straws were filled with soil.
Twist ties were inserted into small holes at the top to create handles that allowed for easy handling of the boxes without disturbing the sampling straws protruding from the sides of the boxes. All rhizoboxes were covered on the sides with black plastic (cut from dog poop bags; top and bottom open) held on by rubber bands to help prevent outside contamination and help with moisture retention in the small boxes, especially once straws were removed for sample collection and open space/exposed roots were present within the rhizobox.
The box was designed so that the top two straws were 1" beneath the soil surface, which is the depth soybeans are planted at, so that at planting the seed simply had to be placed between the top straws in the middle of the box to achieve uniform depth of planting. As the plants grew, the roots would grow straight down between the straws and out into the straws such that soil collected was relevant to where disease would be occurring.
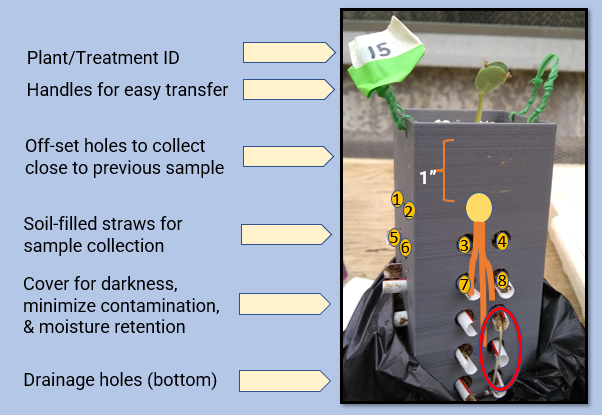
For the field experiment, sheaths were also designed that the rhizoboxes could be securely slipped into to stabilize them and provide space for the protruding straws not to be disturbed. The outer portion of the sheath prevented surrounding field soil from falling into the rhizobox hole, which would have made it challenging to replace the boxes back in the ground after sampling each day. Sheaths were also tagged with labels on twist ties to match rhizobox labels to ensure boxes were returned to their exact place in the field each day. Sheaths had holes in the bottom to ensure drainage. Some design features, like the triangles on the outer part of the sheath, were to minimize cost (calculated based on grams of filament used) while ensuring the sheath could still perform its essential functions and remain sturdy.
Soil Preparation
For the indoor experiments, soil was collected from local fields based on the treatment requirements. The (2) pH treatment soils and the silty clay (proxy control) soil was collected from the Penn State Rock Springs Research Farm. This site did not have sandy soil, which is why for logistical purposes this treatment wasn't included in the field experiment which was exclusively held on the research farm. Sandy soil was collected from a production field in Patton Township, PA.
Soil was collected into plastic tubs from a small area in the middle of the field and was taken from the top 6 inches which is relevant to where seedling growth and Pythium disease would be occurring. Soil cores were collected with a soil probe in a randomized zig-zag pattern within that smaller portion of the field. All four soils underwent testing at the Penn State Agricultural Analytical Services Lab for fertility, OM and N content, and soil particle size testing to provide context for final results and treatment values. Field soil was collected for treatments based on historical tests; however, actual pH levels varied from what was expected. Soil was stored in covered plastic storage bins in a cold room until it was ready to use for experiments.
Soils were sifted to remove large rocks and plant material which could cause sample loss when removing the straw. Soil was then mixed with sand to a volume ratio of 20:80 to prevent compaction in the greenhouse which could influence sample collection and emergence issues unrelated to Pythium. Mixing the soil also made the initial microbiome as uniform in the treatment set boxes as possible. Most soil used in these experiments was unsterilized, but sterile soil for controls in the indoor experiments was autoclaved at 121 degrees Celsius for 30 mins in autoclavable biohazard bags after mixing in the sand. A small amount of bleach-sterilized perlite (<1 cm) was placed on top of the soil (not mixed in) in the indoor experiments after the seeds were planted to protect the topsoil.
No sand was added in the field experiment, as soil from sheath holes was directly placed in the boxes. Care was taken to omit any large rocks or plant material without sifting the soil. No perlite was used, as this would not be used in a regular soybean field production setting. Field sites used were not the same fields from which soil was collected for the indoor experiments.
Inoculation
Soils used in this experiment were collected from field sites with a history of soybean production and were expected to already have Pythium present. However, to ensure pathogen presence, soils were inoculated with Pythium sylvaticum (syn. Globisporangium sylvaticum) which is pathogenic on soybeans. The pathogen isolate originated from a field in Pennsylvania and was obtained from a collection at Franklin & Marshall College, with the species re-verified prior to being shipped for use in these experiments. P. sylvaticum was grown at 25ºC on half-PDA plates containing no antibiotics for four days prior to each experiment. This media was used to minimize its effects on the soil microbiome. Extra plugs were stored in the -80ºC freezer for later use.
Rhizoboxes were inoculated by placing two 5 mm agar plugs 1 inch apart between the top two straws. Non-inoculated soils received media plugs without anything growing on them, in case the media impacted the microbiome. Boxes were topped off with soil and watered. Seeds were planted the next day in between where the plugs were placed between the top two straws by making a small hole with a probe. In the pots, a metal hook was placed into the soil to indicate where the plugs had been placed so that the seed could be set in the middle. In the field rows, plugs were placed about 1 inch apart down the entire row without any indication on their exact location. A hoe was used to make a small furrow for planting the day after inoculation and this inoculated soil was put back on top of the seeds.






Disease Severity Rating
On day 14 plants were harvested and visually assessed for disease symptoms. Rating was done separately for roots and shoots of each plant. Since visual assessment is challenging both a 0-9 and 0-4 scale adapted from the literature and the same single evaluator for each experiment was used to limit bias. As shown in Table 3, root symptoms for visual ratings include germination failure, lesions, brown discoloration, and necrosis, while seedling symptoms focused on brown spots, chlorosis, and wilting or lost leaves.
Table 3. Disease Rating Chart
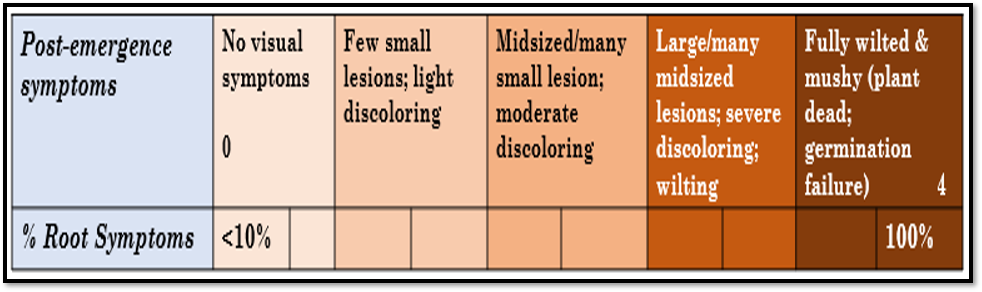
Additionally, notes were made on when plants had emerged during the experiment to identify delays. Photos were taken daily from each set, starting when at least one plant in the treatment/control set emerged, to capture disease development and plant growth. Plant shoot and root lengths in mm were measured, which verifies that the plant roots went deep enough into the box that all straw samples would be relevant. Plants were then cut in half, with the roots and shoots put in separate paper bags labelled by plant number. These bags were put in the greenhouse drying oven for about 2 days before measuring root and shoot dry biomass in grams which might be impacted by disease severity. Plant lengths and biomass were assessed for all indoor experiment plants, both in pots and rhizoboxes. In the field, a representative plant was selected from the middle portion of the numbered row area for assessment.
Disease ratings are linked to the microbiome in analysis and also were used to select soil samples for sequencing. A variety of analyses including significance tests, linear discriminant analysis, correlation and co-occurrence network analysis were conducted using a combination of the open-source software R and Netshift. These analyses are used to identify microbiome communities related to final Pythium disease severity. They also identify highly interactive “hub” microbes and determine their potential for being key microbiome members driving Pythium disease severity during this soybean developmental stage by quantifying directional changes in individual taxa interactions. ROAR, the Penn State Supercomputer, was used to allow the processing of all the microbiome data (over 300 samples) in a single batch within a reasonable amount of time.



Sample Collection
Samples were collected daily over the 14-day experiment timeframe to capture continuous changes in the microbiome during this dynamic soybean growth period and Pythium disease development. Table 4 below describes the soil samples collected for this experiment to support the research objectives. The initial and final timepoints, Days 1 and 14, were hypothesized based on the literature to be the most important timepoints so more samples were collected those days. For the field rows, bulk soil was taken from down the sampling row area (about 1.5 ft between boxes) to create a representative bulk soil sample.
For the indoor experiments, soil samples were collected in the greenhouse facilities. For the field experiments, boxes were brought indoors for straw sample collection in the lab on-site at the farm to limit contamination and for ergonomic reasons. In the first experiment, straws samples were put directly in smaller sterile long-term storage tubes. However, this was very time consuming and the subsequent experiments used longer sterile capped test tubes that the straws could immediately be placed into and later, when convenient, the soil in the straws could be transferred to the smaller tubes. Samples were stored in the -20ºC freezer at the end of each day until use for DNA extraction.
Environmental data, including temperature & relative humidity, were continuously collected throughout the experiment using the Govee and button sensors previously mentioned. Soil moisture was taken prior to watering each day starting with Day 1. The probe was sanitized with ethanol between samples; however, measurements were not taken for any sterile soil controls to ensure they wouldn't get contaminated and it was assumed that each soil type would have similar water retention to their measured non-sterile counterparts. For the pots, probes were set in the center of the pot to collect the soil moisture reading. For the field rows, the probe was set in the center of the row sample site in a similar spot each time.
Table 4. Soil Sampling Overview
| Day | Sample Name | Sample Type | Sample Location | Purpose |
| 0 | Pre-inoculation | Bulk soil |
Taken right before inoculation in that soil location. |
View of the natural microbiome to prior to inoculation. |
| 1 | Post-inoculation | Bulk soil |
Taken from hole between inoculation plugs made directly before seed is placed in it. |
Allows for the verification of inoculation impact on microbiome. |
| 1-14 | Rhizosphere Samples |
Straw samples (Rhizoboxes only) |
1/day/box. Starts with the top straws and zigzags down the box over the experimental timeframe. On Day 1 all boxes were first planted, then straws were collected starting with the first boxes planted. Pots & field rows did not have these samples. |
Understand microbiome community shifts during dynamic plant developmental period & determine best sampling frequency. Determine community shifts and key organisms driving final plant health outcomes. |
| 14 | Root Soil | Bulk soil | Soil adhering to the roots was gently removed at harvest. |
Extra rhizosphere soil sample for comparison to starting samples. This is how rhizosphere samples are collected in destructive experiments. |
| 14 | Bulk Soil | Bulk soil | Soil near the roots in the middle of the box was collected. | Extra bulk soil for more direct comparison to starting samples (which were taken as bulk soil). |
Sequencing & Analysis
Due to resource constraints, a subset of samples expected to be the most valuable in answering the research questions were sequenced. Sequencing focused on the straw rhizobox samples for comparability and only non-sterile samples were sequenced since we want to track the natural microbiome under Pythium disease pressure. One plant of each treatment set was not sequenced at all and stored for back-up purposes. Plants included for sequencing were selected using standardized selection parameters across treatments based on experiment plant growth and disease severity.
All plants were sequenced for days 1, 7, & 14, which were hypothesized to be the most meaningful timepoints. Seven (7) days is about when emergence occurs for soybeans. All remaining samples are kept in -20ºC freezer storage in case they are needed for future research or analysis building off this project. Table 5 below shows the specific days each plant in a treatment had sequenced.
Table 5. Sampling Scheme for Each Experiment
| Plant Replicate | Sequenced Sample Day |
| Plant 1 | 1 - 14 |
| Plant 2 | 1, 3, 5, 7, 9, 11, & 14 |
| Plant 3 | 1, 7, & 14 |
DNA extraction of (4) plates of soil samples was done using the vacuum pressure protocol of the Macherey-Nagel NucleoSpin 96-well Soil DNA Extraction Kit. Extract plates were sent to the University of Michigan Microbiome Core Facility, which offers more full-service sample preparation at a reasonable price per sample, for MiSeq sequencing of the 16S (bacteria) and ITS (fungi) regions. Samples with poor MiSeq results were removed prior to data processing. Remaining MiSeq results were processed using the dada2 pipeline in an interactive R Studio interface on the Penn State ROAR Supercomputer, which allowed the processing of all 300+ samples together in a reasonable amount of time. Various analysis such as Shannon diversity indices and relative abundance was used to visualize soil microbiome communities. Principal components analysis visualizes differences in communities based on environmental and soil characteristics, while principal coordinates analysis and significance tests highlight differences between treatments. Altogether, this genomics data was used to visualize the abundances and diversity of fungal and bacterial microbiome community members throughout this early soybean development period under Pythium disease pressure.
Soil Characteristics
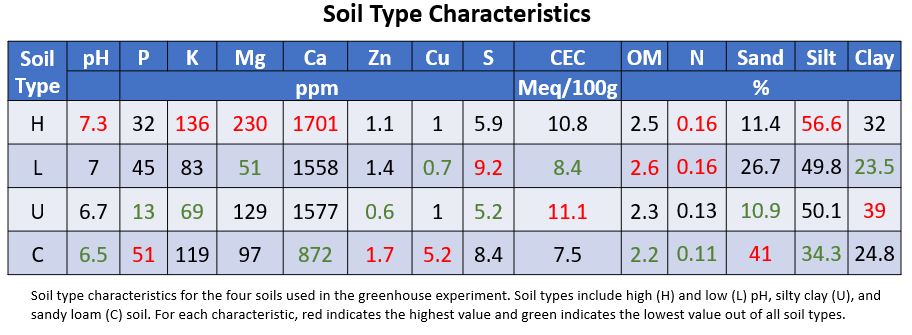
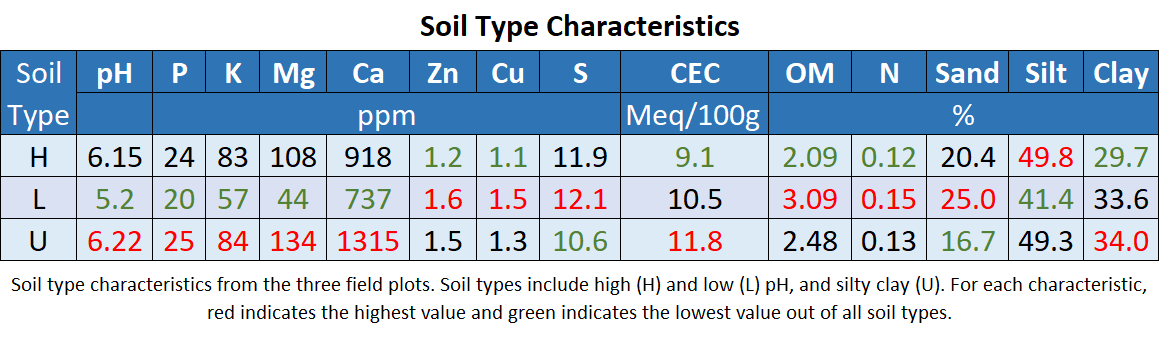
The high and low pH soils used in all experiments were selected based on expected characteristics from previous soils tests, as the results from the soils sent to the lab were not received until after the experiments began. Although ultimately there was a well-scaled pH range of 6.5 to 7.3 across all four soils used in the indoor experiments, the treatment labeled as low pH had a neutral pH and the sandy loam used had the lowest pH. Based on the fertility tests, the sandy loam's pH of 6.5 is below optimum.
All experiment soils had levels of the trace element sulfur below the normal range found in PA soils. In the indoor experiments, all but the sandy loam had less copper than is typically found in PA soils. The low pH soil had both magnesium and potassium levels below optimum. Both the high pH soil and silty clay soil had magnesium levels above optimum, while the sandy loam had phosphorus levels above optimum. The primary soil used in the experiment had low phosphorus, potassium and zinc.
For the field experiment, there was no sandy treatment and soil in boxes was used directly from the plot sites. The silty clay soil (U) used as the control soil in the experiments had a pH similar to the high pH soil. All pH's, phosphorus ppm, and potassium ppm were below optimum levels, while all magnesium levels were optimal or above. Zinc and copper were in the expected range for PA soils. The levels of some of these nutrients outside their optimal or typical range could explain some of the symptoms seen in the experiment across treatments.
Disease Ratings
In general, disease ratings were high indicating lower plant health across all experiments and treatments. The soil used was not known to be suppressive of Pythium disease and these high ratings suggest a successful Pythium inoculation. Ratings were in alignment with what was expected for that environmental characteristic. For example, saturated soil had high disease and water is important for Pythium infection and motility. Silty soil, which has better water retention, had higher disease ratings than sandy soil. The fungicide varied in efficacy, which is what farmers experience in the field, but did appear to slightly reduce disease severity. There was no clear pH impact on disease, which may be due to pH influencing a range of soil abiotic and biotic factors that influence Pythium disease differently.
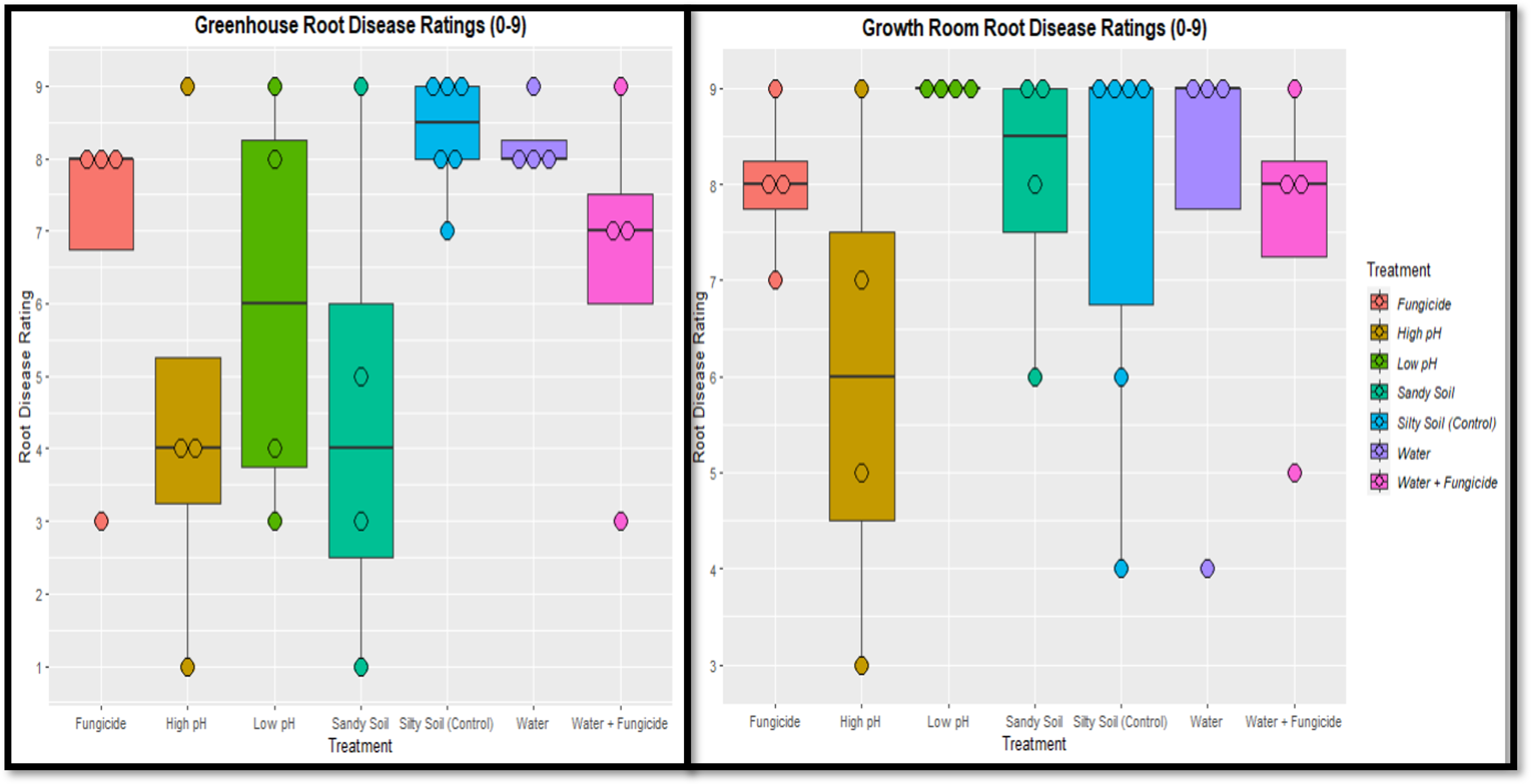
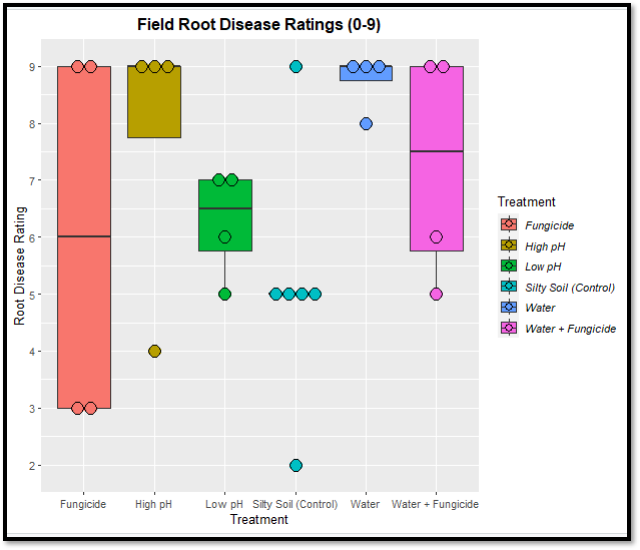
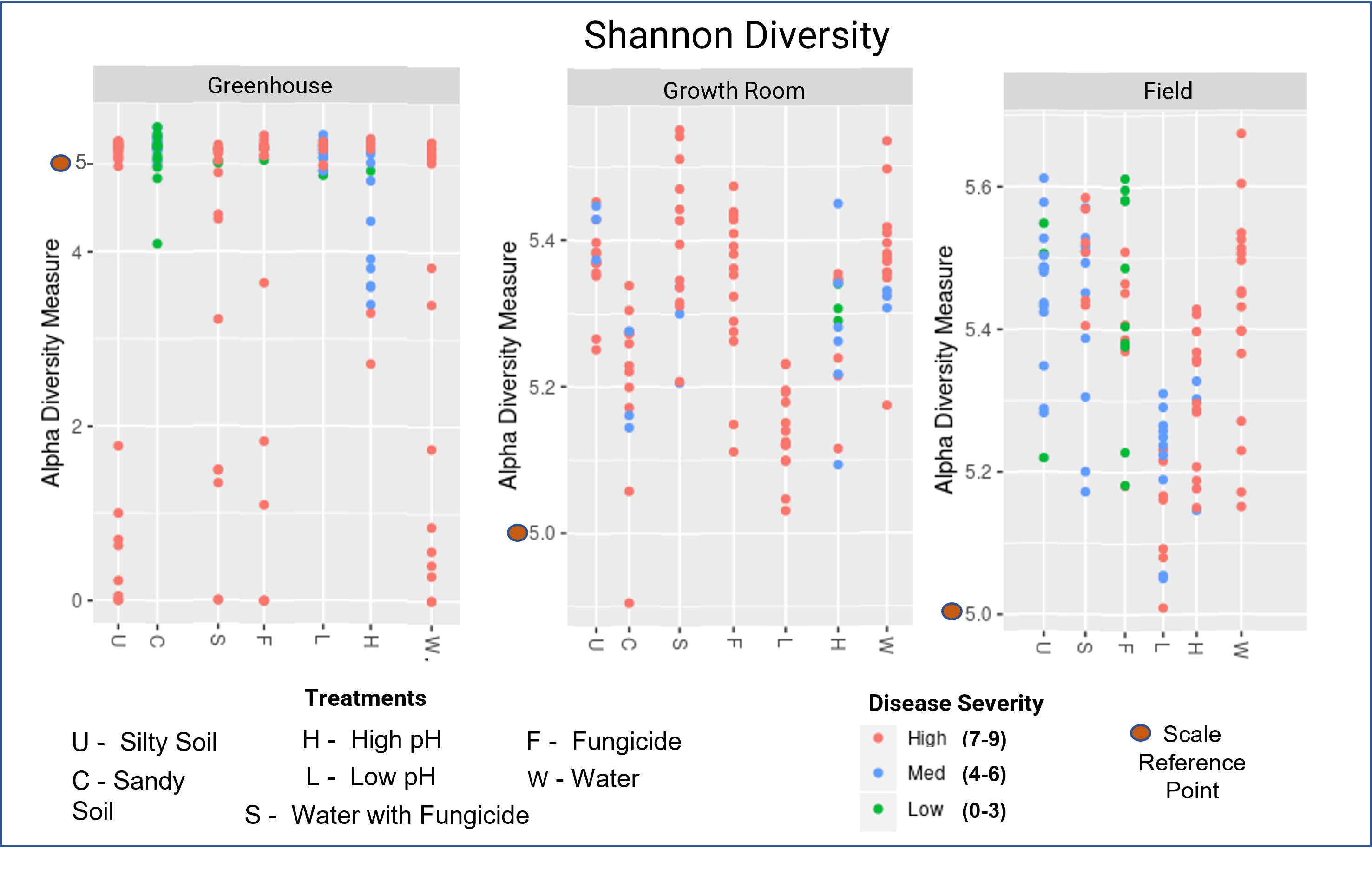
Diversity & Abundance
There was high bacterial diversity seen across all treatments and experiments, predominately with a Shannon index value ranging from 5.0 to 5.5. High diversity is expected in soil samples from a natural setting. There were some low diversity samples seen in the greenhouse experiment and all of these samples had a high disease rating. Overall, low and high disease ratings were intermixed within the same diversity range suggesting that bacterial diversity in general is not predictive of disease outcomes.
Acidobacteriota and Proteobacteria were the most abundant bacterial phyla. Acidobacteriota and Verrucomicrobia, another abundant phylum, have been associated in other soil plant pathogen systems with diseased plants. Furthermore, the abundance of actinobacteria which has been correlated to increased soil suppressiveness of plant pathogens in other studies was smaller compared to the other disease-associated phyla. These results are logical considering the high disease ratings seen in these experiments; clearly Pythium suppressive soils were not used. Altogether, these results suggest that the abundance of these phyla could help predict plant health across systems. Further research on specific organisms at lower taxonomic levels in these phyla can provide options for developing tools for disease management decision-making and new biocontrol options for integrated pest management against Pythium and other diseases.
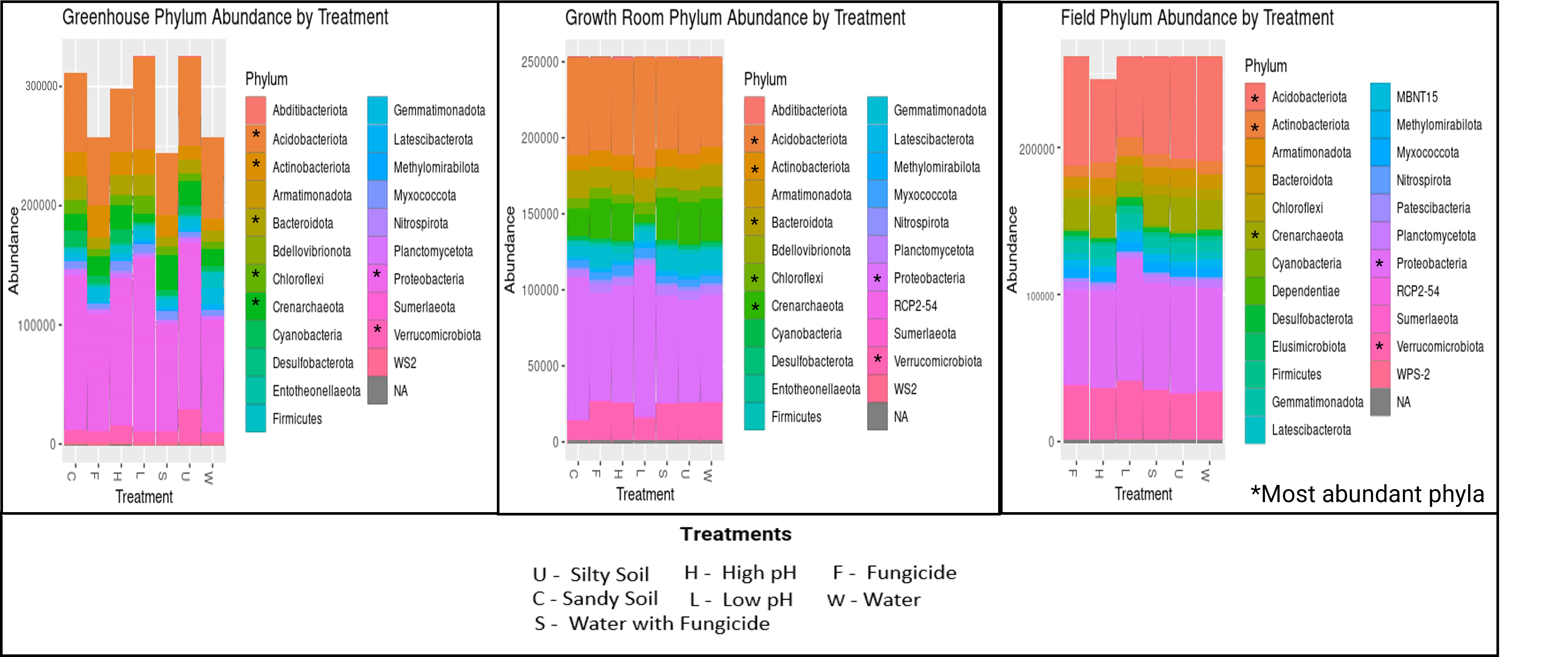
In summary, environmental factors considered in this study (pH, soil type, fungicide, and soil moisture) did not drive dramatic shifts in the soil microbiome during this 14-day early soybean development stage under Pythium disease pressure. Despite significant changes in plant development, this timeframe may be too short for large-scale soil microbiome shifts that can drive Pythium disease outcomes. The majority of plants rated medium to high disease severity, which indicates we did not use Pythium suppressive soil in this experiment. Since the abundance and diversity of the soil microbiome did not dramatically shift over the 14-day early soybean development timeframe, the initial microbiome would be a suitable timepoint for disease management decisions. This suggests that farmers need to have an established Pythium suppressive microbiome prior to planting, which would require soil management decisions in advance of the growing season. This research also highlights the variable efficacy of fungicides which supports farmers decreasing fungicide use in favor of more sustainable alternatives that could offer more reliable results.
In particular, key bacterial phyla like Acidobacteriota and Verrucomicrobia that have been associated with diseased plants in other plant and pathogen systems were found in this study in high abundance. Their presence in the microbiome at the time of planting can inform farmer's expectations of disease outcomes that drive their management decisions. Future research can focus on how to reduce the abundance of these disease-associated phyla to enhance soil suppressiveness and elucidate specific organisms in these phyla that can be targets for the indirect control of Pythium. Developing suppressive soils will provide a long-term sustainable defense against a variety of soilborne plant pathogens that will support farmers into the future.
Education & Outreach Activities and Participation Summary
Participation Summary:
A major goal of this project is to provide the scientific community with fundamental knowledge of how the soil microbiome communities of fungi and bacteria influence plant health outcomes during the early soybean development stage. The results support future in-depth studies of key driver organisms to understand biocontrol mechanisms and develop management strategies targeting disease-driving organisms.
This research has been presented across the United States (see table below), as the knowledge gained is not limited in value to only Northeastern agriculture. A short demonstration of the field experiment was provided to the PI's lab group in May 2022. Additional publications, educational materials, and outreach are expected to continue to develop from this project. These educational materials will include photos and videos collected from the experiments to illustrate various aspects of the project.
| Event | Location | Estimated Participants | Timing |
| Plant Pathology & Environmental Microbiology Departmental Lecture Series |
University Park, PA The Pennsylvania State University In-person & Zoom |
30 students/faculty/researchers/public |
April 2021 |
| Northeastern American Phytopathological Society Meeting |
Salisbury, CT Heritage Hotel In-person only |
50 students/faculty/researchers/extension specialists |
March 2023 |
| Conference on Soilborne Plant Pathogens |
Salinas, CA Monterey County University of California Cooperative Extension Office In-person only |
100 students/faculty/researchers/extension specialists/industry |
March 2023 |
|
Annual American Phytopathological Society Meeting Published Abstract Submitted March 2023 |
Denver, CO Sheraton Downtown In-person only
|
100 students/faculty/researchers/extension specialists/industry |
August 2023 |
Project Outcomes
This project highlights how the microbiome bacterial and fungal communities can help explain differences in disease outcomes seen under fungicide application or within a field under similar environmental conditions. Abundances of driver taxa may be a useful indicator for predicting plant health to help inform a farmer's management decisions and minimize unnecessary interventions that affect farmer profit or drive pathogen resistance. Alternative sustainable management strategies based on driver taxa identified in this project such as biocontrol development or suppressive soil cultivation will require time to develop, test and be approved for use, but will benefit future agricultural sustainability through long-term disease suppression with limited inputs. The availability of sustainable alternatives to chemical controls will lead to future reduction of chemical applications in an integrated pest management plan leading to wide-spread benefits to environmental and human health. Supporting higher crop yields sustainably and without expensive chemicals will provide economic benefits to farmers, which may be passed on to consumers, leading to additional social benefits.
This project increased my understanding of the skills required for and challenges involved with microbiome research that supports sustainable agriculture. It has also raised awareness of the complexities involved in developing this knowledge into an applicable tool for farmers. Farmers should be included throughout the process, such as by conducting sustainable agriculture studies via on-farm research programs, to promote stakeholder buy-in for implementing sustainable agriculture practices. I acknowledge that research to support sustainable agriculture requires a collaborative multi- and inter-disciplinary approach and a dedicated team invested long-term in developing new effective disease management strategies that will withstand changes to the agricultural system through time.
This project has generated new research ideas that I can study further during my career to support agricultural systems and their adaptability to social and environmental changes. It has allowed me to develop highly transferrable knowledge and skills which will be invaluable throughout my career. I plan to pursue interdisciplinary research projects because I enjoy exploring different perspectives and approaches addressing the same issues and integrating them to develop unique solutions. I am particularly interested in working at the nexus of crop and animal agriculture, with a focus on food safety and security under sustainable agriculture production. In addition to the agricultural research side, I am also interested in the product marketing, regulation, and consumer end of production. I want to be involved in science communication and outreach with farmers and the public, as well as be involved in the development of science policy that supports sustainable agriculture to enhance the impact of this research area.
3D Printing
This allowed customization of a box highly specified to my needs and the material used was biodegradable. I would have preferred clear material so I could visualize root growth throughout the experiment for educational purposes, but this was not available at my 3D printing facility. Additionally, I wanted to develop a 3D printed probe to use instead of the straws but ran out of time before I had to start the first experiment. Boxes can't be autoclaved, which may be an issue for some other projects, but bleaching them before the experiment seemed sufficient for our sterilization purposes.
Another benefit of 3D printing is that the file sent to printing can be shared by other researchers for reproducibility in their own projects. My impression is that 3D printing is seen as inaccessible or difficult by agricultural scientists and in my research presentations I work to highlight how easy it really is to make objects for printing to change that mindset. 3D printing spaces are becoming more readily available in communities and could be a valuable tool for future research on sustainable agriculture.
Rhizoboxes
The rhizobox sampling method worked well overall; in some cases roots were growing directly in the sampling straw so one knew that soil collected was relevant to where the disease would be occurring. Watering had to be done slowly to prevent soil from being pushed out of the box, but excess soil loss from the boxes wasn't much of a problem as long as care was taken when handling them. After straws were removed for sample collection, soil didn't necessarily fall down into the open space, such that you could see through the box, which potentially could have influenced plant growth or outside microbial contamination. Boxes were covered with plastic bags to help with water retention and to minimize contamination, but as the plants grew taller sometimes when removing the rubber band holding them on leaves were lost which affects disease ratings and biomass data at the end of the experiment. Designing a bottom slip-on 3D printed cover would have helped with this, but plants were not expected to get quite as tall as they did in the first experiment. Root biomass loss may have occurred when extracting plants at harvest from the boxes as it is challenging to ease them out of the boxes and soil without losing some small pieces, but this is not an issue unique to the rhizoboxes.
Future researchers can build off the rhizobox design from this project and tailor it for their own needs; for example, many aspects like the spacing, hole and box size were specifically used to account for expected soybean growth during the experiment and other unique needs of this project and crop. It also takes printing several prototypes to finalize a design and hours to print so box preparation should occur well in advance of when they are needed.
Sample Collection
The straws were effective for sample collection to obtain a sufficient amount of soil from the relevant soil location of where the plant is growing; however, their preparation prior to the experiment was tedious and time-consuming. It also took a bit of time to transfer the straw samples to the freezer storage tube; therefore, in 2022 straws were put directly into sterile test tubes and the soil was transferred when convenient in the lab to the smaller long-term storage tubes. Since transfer was done in the greenhouse the first year, there were more opportunities for external contamination of samples and the heat could have affected the microbiome over the time it took for collection. The test tubes allowed faster and more consistent sampling times on experiment days.
Inoculation & Disease Rating
Creating a liquid inoculum of Pythium zoospores following protocols in the literature was not successful so the agar plug inoculation method was used in this experiment. The efficacy of this inoculation was not verified outside of the microbiome sequencing data; this experiment did not reisolate Pythium from plants or verify that disease symptoms were specifically caused by Pythium. For example, May 2021 was abnormally hot in Pennsylvania and the plant shoots generally appeared to grow quite tall and healthy-looking. Plants primarily exhibited leaf symptoms, such as chlorosis and brown spots, which were not necessarily related to Pythium disease and may have been caused by nutrient deficiencies which can occur in greenhouse trials. Since Pythium prefers cooler environments, the following year we moved to a walk-in growth room in the Penn State Greenhouse Complex to allow better temperature control. More symptoms expected of Pythium like delayed or no emergence and clearer root rot were seen in the 2022 experiments.
Root disease severity is challenging to visually rate and both a 0-4 & 0-9 rating scale were used, with the 0-4 scale being easier to use. Ratings were done by the same one person each time to try to limit the effect of natural biases. Future researchers can consider using software to quantify disease severity and further limit bias for more confidence in visual disease ratings. Additionally, they can consider a more holistic plant heath score that combines visual ratings with physical characteristics like shoot & root biomass or length.
Other Challenges
Slugs
Slugs were an issue in the field experiment. They were seen moving inside and on the outside of the boxes, found eating the soybean plants, which affects final shoot data, and laying eggs in the straws. Their eggs could have accidently contaminated soil samples used for DNA extraction and affected microbiome analysis results. I would recommend these boxes for slug research or scouting purposes.
Scaling
Particularly for the first greenhouse experiment scaling issues occurred. Additional plants in pots had been added to the original design since not all plants grew during the rhizobox design tests, which was expected due to germination failure and disease, to ensure enough post-emergence disease data would be available. However, pandemic protocols made it challenging to obtain multiple dedicated project assistants and the scaling made it very intensive for one person to complete the experiment. In the end, almost all plants emerged (over 140) which led to significantly more effort required at harvest than was expected. Since extra plants and an extra experiment were added, I had to be more selective of what samples were preliminary sequenced. Altogether, several recommendations for future researchers would be to confirm assistants dedicated to the project well in advance and obtain feedback on changes to the number of experimental plants to verify resources. Additionally, finding additional funding for microbiome analysis when scaling up an experiment can allow one to answer more research questions that evolve out of a project.
Soils
In general, soils used for this experiment were selected based on expected pH from farm data which was verified by soil fertility testing after the experiments started at the Penn State Agricultural Analytical Services Lab. For this reason, pH levels used in this project were different than expected for the treatments, but still varied enough for a soil pH treatment comparison. This delay in soil analysis was so that soil for the first greenhouse experiment wasn't collected too far in advance to minimize storage and hopefully preserve soil microbial communities as close as possible to the original microbiome at the time the soil was collected from the field. Soil samples tested were representative of the small area of the field where the experiment soil was obtained to ensure it was relevant for abiotic treatment analysis and field sites were only able to be set up so far in advance. Soil collected for, but not used in, the 2021 experiment was saved in a cold room for the 2022 experiment and it would be interesting to see if results would have differed if we collected fresh soil from the fields in 2022.