Final report for GNE21-267
Project Information
Bacterial blotch refers to a group of diseases characterized by the formation of spots and discoloration on the mushroom cap. Blotch diseases are common threats in mushroom production and were once thought to have been caused by a few Pseudomonas species. Several non-Pseudomonads spp., including Mycetocola, Burkholderia cepacia, and Ewingella americana have been identified as causal agents of bacterial blotch and found in different combinations with other species on the surface of mushroom caps. The complete diversity of pathogens causing blotch on white button mushroom caps is still unknown. One barrier to discovering the complete diversity of blotch diseases is the labor-intensive nature of pathogen isolation and identification. Using traditional low-throughput schemes, bacteria are isolated on non- or semi-selective media followed by multiple rounds of purification and storage of individual isolates prior to intensive pathogenicity testing. This results in a high demand for supplies, hands-on effort, time, and space to conduct the study. In this study, a high-throughput bacterial isolation system and traditional isolation scheme were used to recover pathogenic bacteria from symptomatic mushrooms. The Prospector System ™ (manufactured by General Automation Laboratory Technologies (GALT)) was used for high-throughput isolation, cultivation, and screening of bacteria from symptomatic mushrooms in Pennsylvania. High-throughput isolation and standard isolation, followed by pathogenicity tests, were used to build a collection of blotch pathogens. Species identification was determined by amplifying the rpoD gene. Using both the low- and high-throughput schemes, four distinct clades of blotch pathogens were successfully recovered in Pennsylvania and 16 distinct clades were recovered from the California mushroom facilities. Our results indicate high-throughput technology has the potential to be a useful tool to help enhance our understanding of the spectrum of blotch pathogens present in mushroom production.
Aim 1: Determine whether non-Pseudomonas species are responsible for causing bacterial blotch in mushroom production in Pennsylvania using two distinct approaches. The first approach will use a high-throughput system to exhaustively isolate non-fluorescent organisms from a limited number of symptomatic mushrooms. The second more traditional approach, will be labor-intensive and isolate a few individual organisms from many different symptomatic mushrooms. Non-fluorescent isolates will be tested for pathogenicity using Koch’s Postulates.
Aim 2: Determine the identity of the non-Pseudomonas blotch pathogens from aim 1. 16S rDNA sequence analysis will provide preliminary identification of the pathogenic organisms isolated in aim 1. If appropriate, other identification techniques such as MLSA or phenotypic tests will be used to confirm the identity of organisms.
Cultivated edible mushrooms have been on the rise since the late 1800s (Royse et al., 2017). Agaricus bisporus, or the button mushroom, is one of the most common edible mushrooms produced and consumed in the US (Robinson et al., 2018). Between 2019-2020, Agaricus crop production valued at an estimated $1.15 billion US dollars. In the US, Pennsylvania is responsible for leading the country in button mushroom production (USDA, 2020). Bacterial blotch is a complex of diseases threatening the mushroom industry, resulting in discoloration and deformation of the mushroom cap (Tolaas, 1915). Blotch diseases can lead to severe economic loss by reducing the marketability of the crop by lowering the expected shelf life after harvesting, reducing the standard of the product, and reducing crop yield (Navarro et al., 2018). Diverse species of Pseudomonas have been identified as being the causal agent of blotch diseases in mushroom production (Martins et al., 2020). In 2016, the mushroom industry identified bacterial blotch as one of the most important diseases of mushrooms during a Penn State University Industry Strategic Planning meeting. Management of blotch caused by Pseudomonas became one of the primary goals of a subsequent USDA OREI grant.
Bacterial blotch results in the formation of brown spots and sunken lesions on the surface of mushroom caps (Munsch & Alatossava, 2002). As part of the OREI project described above, Martins and Bull identified 10 distinct species of Pseudomonas that cause blotch in Pennsylvania (Martins & Bull, 2019). Similar results were found in Europe (Taparia et al., 2020), all shown to vary in symptoms and disease progression. P. agarici can cause drippy gills or yellow blotch on the mushroom cap (Young, 1970), while P. costantinii and P. tolaasii have been shown to cause brown blotching on the mushroom cap, and P. gingeri have been shown to cause discoloration on the mushroom cap (Osdaghi et al., 2019). These findings are just a couple of examples that highlight the great level of diversity within Pseudomonads that cause bacterial blotch. In recent collaboration between Hamidizade and our team, novel non-Pseudomonads pathogens from Iran were identified but currently no similar study has been conducted in the US (Hamidizade et al., 2020). The spectrum of blotch pathogens causing this disease in US mushroom growing regions is still something that needs to be explored. Understanding which blotch pathogens are responsible for causing this disease will help in targeting the spectrum of pathogens responsible in future management practices.
Research
Aim 1
Sample Collection
Diseased mushrooms were collected from two commercial mushroom houses in Pennsylvania - Marlboro Mushrooms, Phillips Mushrooms. Two mushrooms per facility will be collected for deep sampling using the Prospector System ™ (approach A). In addition, one hundred mushrooms were collected for traditional isolation (approach B) from each facility. Each mushroom cap was stored in a sterile specimen cup in a cooler and transported from facilities in Chester or Burks County to the Penn State University Park campus for processing.
Approach A – High-throughput deep sampling
I employed the unique features of the Prospector System ™, a high-throughput cultivation platform, to isolate non-fluorescent organisms present in the symptomatic tissue. The Prospector System ™ allows for the isolation of up to a thousand colonies from each sample with minimal manual labor. The mushroom lesions were cut, surface sterilized, macerated in buffer, and passed through a filter to remove lingering mushroom tissues from the sample. The sample was then inserted into the Prospector System ™ array where individual cells will grow in isolation. Pseudomonas blotch pathogens fluoresce in King’s Medium B (KMB) while no other bacteria are fluorescent in this medium (King et al., 1954). The Prospector System™ was programmed to select non-fluorescent isolates and transfer them onto a 96 well plate for long-term storage and subsequent pathogenicity testing. No additional labor was required after the initial sample preparation in order to obtain large numbers of purified cultures of non-fluorescent isolates from symptomatic mushrooms. Preliminary data indicated an average of 27 non-fluorescent isolates were recovered from each sample using this approach (Herschlag et al., unpublished results).
The Prospector System ™ is designed to enhance the process of studying microbial organisms by scaling up the number of isolates retrieved within a given sample compared to traditional approaches. This technology has never been used to study the diversity of blotch causing pathogens, symptomatic mushrooms were used to troubleshoot the system’s ability to select fluorescent and non-fluorescent organisms. Seven mushrooms were collected August 2021 from three different mushroom facilities in CA – Royal Oaks Mushroom Farm, South Valley Mushroom Farm, and Morgan Hill Mushroom Farm. From this sampling a total of 870 isolates were retrieved, 684 selected to be fluorescent and 186 selected to be non-fluorescent organisms as shown in table 1.
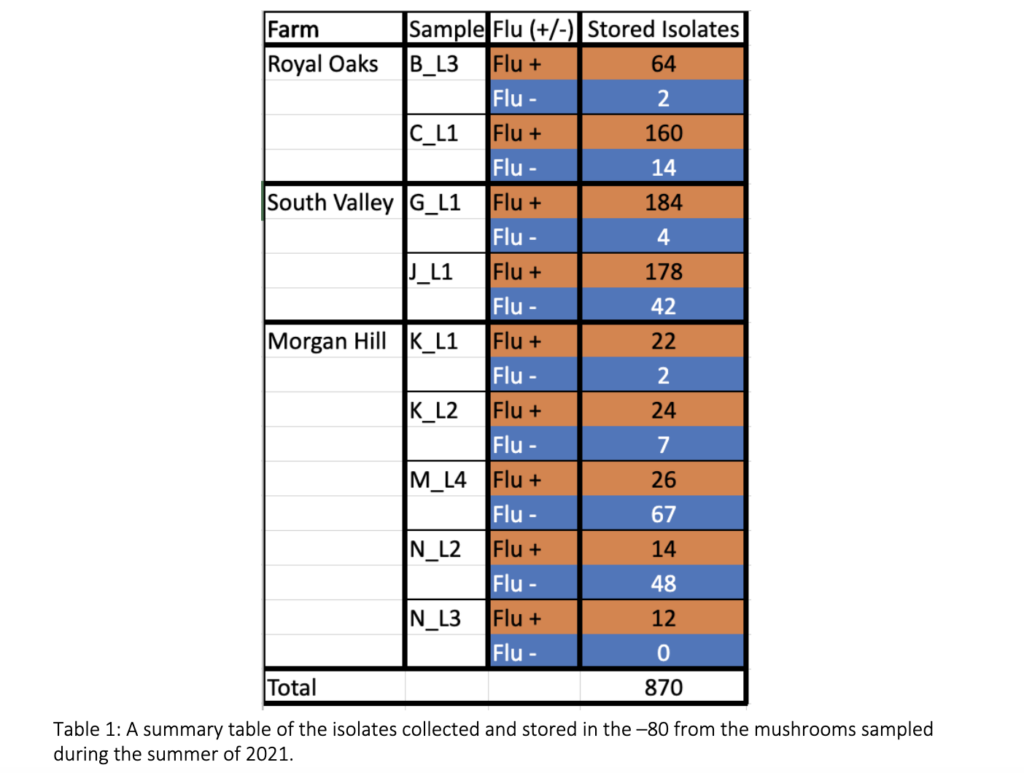
Approach B – Traditional sampling
An additional 100 mushrooms from each of the three organic mushroom facilities were sampled. The lesions from the diseased mushrooms were removed, surface sterilized, macerated in buffer, and the bacterial sample were streaked for single colonies on KBM with cycloheximide to screen for non-fluorescent colonies. Pure cultures were generated by multiple rounds of isolation, starting with single colonies. Each isolate was stored at -80°C in 50% nutrient broth 50% glycerol stocks. This approach is labor-intensive and would require inoculating at least 300 agar plates for processing the samples from a single facility. Likewise, to test for pathogenicity, isolates must be grown on individual petri dishes for one or more rounds to prepare inoculum.
48 isolates were collected from the five mushrooms collected from Marlboro Mushrooms. From this sample, 40 fluorescent and 8 non-fluorescent isolates were identified. Phillips Mushrooms and an additional mushroom farm identified by AMI will be used to collect future samples.
Pathogenicity Testing
To test bacterial isolates for pathogenicity, 10 µl of bacterial suspensions were spotted onto surface-sterilized mushroom caps where the surface had been removed with a sterile knife. Mushrooms were incubated in a moisture chamber for a 72-hour period with observations recorded every 24 hrs. A disease scale was developed by documenting symptoms caused by known blotch pathogens from a current strain collection and used to rank the level of disease severity by blotch pathogens. Pathogenicity was confirmed via Koch’s Postulates.
One potential pitfall would be to not find non-Pseudomonas pathogens from these facilities, if that is the case mushrooms will be sampled from additional facilities.
Aim 2
Taxonomic characterization
16S rDNA sequence analysis was used for the preliminary identification of the isolates. The 16S rDNA for each non-fluorescent mushroom pathogen was amplified using universal primers for 16S rDNA (Weisburg et al., 1991). Amplicons were Sanger sequenced at the Penn State Genomics Core facility. 16S sequences were compared to previously identified Pseudomonas blotch pathogens.
Pseudomonas species identification was conducted by amplifying the rpoD gene. It has been demonstrated that the amplification of housekeeping genes, such as rpoD, can be used to distinguish Pseudomonas species down to the species level (Yamamoto et al., 2000; Liu et al., 202; Martins, et al., in prep). Sanger sequencing of the amplicons were conducted at the Penn State Genomics Core facility and rpoD sequences were extracted from Hesse et al. 2018, Girard et al. 2021, Laucat et al., 2022 and from blotch pathogens (Taparia et al., 2020; Martins et al., in prep; Liu et al., 2022). To generate neighbor joining trees, the sequences were evaluated, aligned, and trimmed using CLC workbench. To identify organisms down to the species level, pairwise comparisons were generated using a cut-off value of 98% nucleotide identity (Girard et al. 2020).
500 isolates were recovered using the high-throughput approach and 10 isolates recovered using the low-throughput approach from the same two mushrooms collected from Pennsylvania mushroom production (Fig. 1). These findings indicate both methods successfully recovered four distinct species of blotch pathogens. Fig 2 shows the sequencing results of the 615 isolates recovered using the high-throughput approach in California mushroom production out of the seven mushrooms that were sampled from three different farms, 16 different clades of blotched pathogens were recovered. Fig. 3 shows three representative strains from each group from the California and Pennsylvania survey, because the isolates recovered in Pennsylvania resulted in four distinct species and the isolates recovered in California resulted in 16 distinct species.
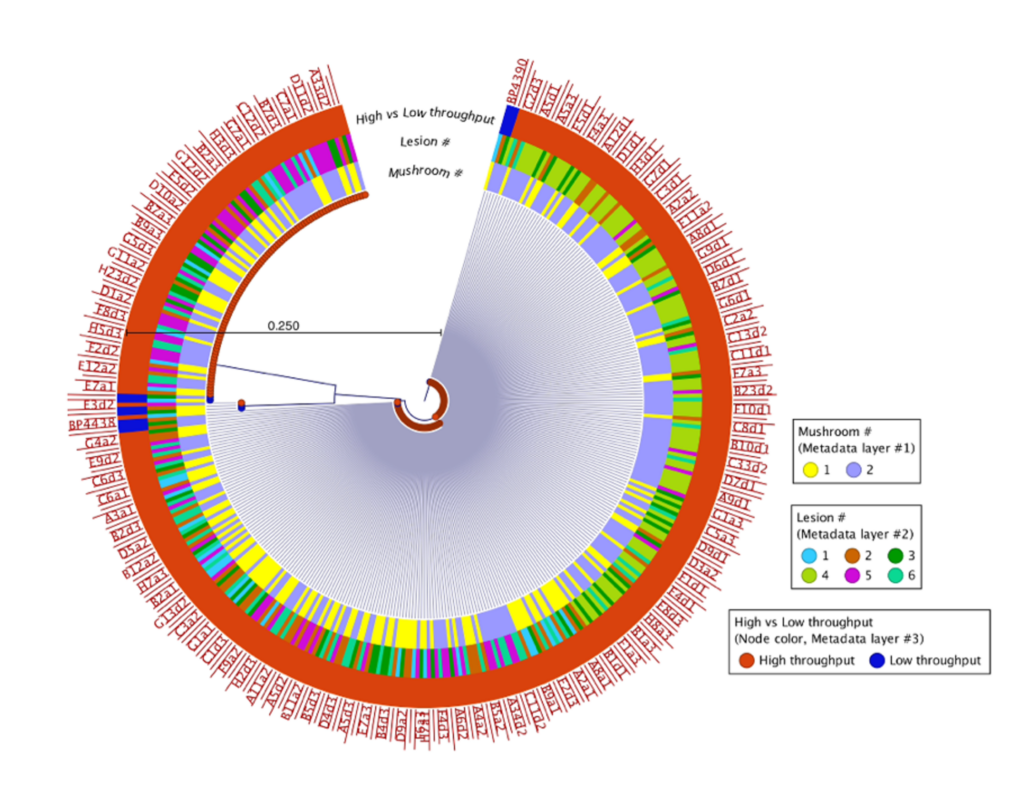
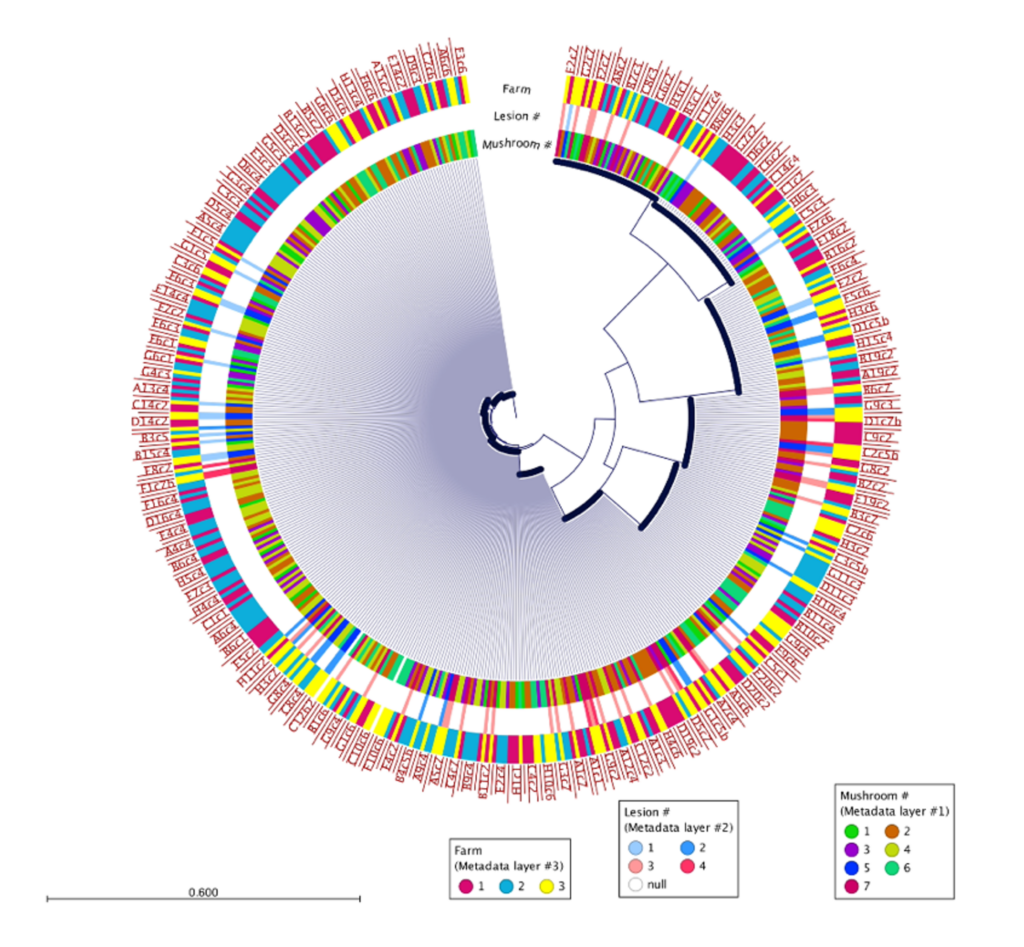
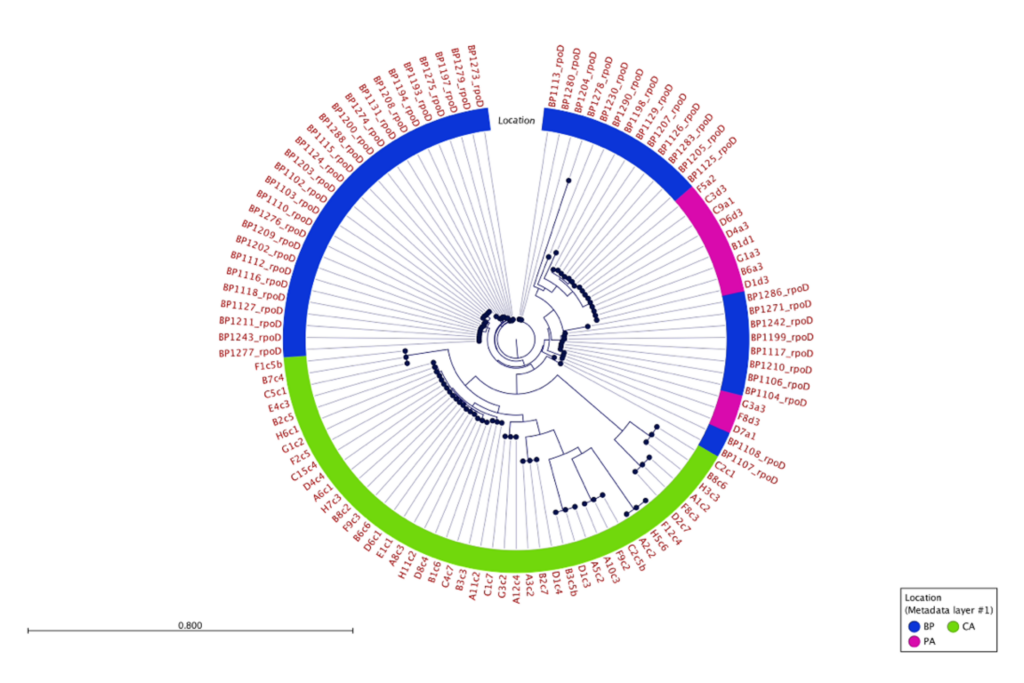
In the Pennsylvania survey, four distinct species of blotch pathogens were successfully recovered using both the low- and high-throughput approach. In this survey, the number of isolates that were recovered using the high-throughput approach were scaled up in comparison to the number of isolates recovered using the low-throughput approach. Given that the same mushrooms were used in this survey for both high- and low-throughput isolation approaches, it is not completely surprising that both approaches recovered the same clades of pathogens. The difference in diversity between the two surveys might indicate that increasing the number of mushrooms and farms sampled will recover greater pathogen diversity, or that there is generally higher pathogen diversity in California mushroom houses compared to Pennsylvania. The Pennsylvania and California survey results suggest that future surveys should consider diversifying the mushroom used for future studies. Importantly, the results of this work demonstrate that the clades of pathogens in Pennsylvania are distinct from those in California, indicating a strong geographic influence on pathogen populations. Moving forward the Prospector System ™ has great potential to be used in exploring the diversity of blotched pathogens. Future studies that implement high-throughput technology could help enhance our understanding of the spectrum of blotch pathogens present in additional mushroom production regions and provide insight on how to develop specific biocontrol strategies targeted at these pathogens.
Education & Outreach Activities and Participation Summary
Participation Summary:
Extension
The mushroom industry is currently not using the Prospector System ™. The first presentation presented the potential to use this approach for their industry. Research findings were presented to mushroom growers and other industry professionals as they become available. Oral presentations were presented at the 62nd & 63rd annual Penn State Mushroom Short Course meeting September 2021 and 2022. The Mushroom Short Course is an annual event designed to connect mushroom growers to current research addressing issues threatening mushroom production. An oral presentation was delivered in September 2021 to the International Society for Mushroom Science (ISMS) which is an industry-run conference designed to connect growers on an international level (Richardson et al., 2021). Lastly, the completed work was presented during the Plant Health 2022 annual meeting to members in the American Phytopathological Society.
Project Outcomes
The results from the sampling conducted in CA indicate the Prospector System ™ has the potential to be a useful tool for studying the diversity of blotch causing pathogens by selecting a large set of isolates compared to traditional approaches and enhancing the diversity of blotch pathogens captured within a given sample. Incorporating this technology into the mushroom industry could enhance the grower's knowledge of the pathogens present in their system.
814 of the isolates processed using the Prospector System ™ were submitted to be Sanger sequenced at the Penn State Genomic Core facility. Using both the low- and high-throughput schemes, four distinct clades of blotch pathogens were successfully recovered in Pennsylvania and 16 distinct clades were recovered from the California mushroom facilities. Our results indicate high-throughput technology has the potential to be a useful tool to help enhance our understanding of the spectrum of blotch pathogens present in mushroom production.
In addition to the identification of blotch pathogens using high-throughput isolation techniques, I along with a Penn State undergraduate Millennium Science Scholar have worked on assessing the host range of known blotch pathogens of Agaricus bisporus on a variety of exotic mushrooms. We evaluated the virulence of 11 species of Pseudomonas blotch pathogens previously demonstrated to cause bacterial blotch disease on white button mushrooms grown in Pennsylvania by the Bull lab.
Moving forward the Prospector System ™ has great potential to be used in exploring the diversity of blotched pathogens. Future studies that implement high-throughput technology could help enhance our understanding of the spectrum of blotch pathogens present in additional mushroom production regions. The results from this study suggest there could be a spectrum of pathogens responsible for causing bacterial blotch, this information could provide insight into the development of specific, sustainable, biocontrol strategies targeted at these pathogens present in the mushroom cultivation system. The future direction for my career would be to pursuing research that would allow me to utilize natural microbial systems to improve industrial process while reducing the downstream impacts from industry pollution.