Final report for GNE21-269
Project Information
The problems to be addressed
Urban agriculture (UA) holds significant potential to feed the growing urban population and offers numerous environmental and societal advantages for urban areas. However, poor soil health with low soil organic carbon (SOC) is one of the major concerns preventing high yields and profits in UA. In UA, many farmers generate their growing soil media by decomposing their farm biowaste, local green waste, or food waste into organic matter via a composting process. However, due to the nature of this type of artificially created soil, many urban composts lack a good physical structure, soil microbial activity, and nutrient availability. In addition, the nutrients in urban composts may not be properly retained within the soil environment and are prone to leaching into the urban groundwater system, causing significant environmental issues such as eutrophication. Despite its importance, urban gardening soil has largely been neglected by the research on UA. The ecological interactions that support urban soil's biochemical and ecological processes, which are essential for effectively regulating soil health in UA, are only partially understood.
The solution to be pursued
To address the long-standing issues that urban farmers currently experience, we aimed to reveal the ecological functions of soil microbiomes to enhance plant performance and soil health in UA. In the context of conventional field-based farms, beneficial soil microbes have garnered significant attention in terms of improving soil quality and plant productivity in organic agriculture. These include microorganisms such as phosphorus-solubilizing bacteria (PSB) and arbuscular mycorrhizal fungi (AMF), owing to their ability to enhance soil nutrient cycling, plant nutrient uptake, plant growth, plant disease resistance, soil aggregation, and SOC sequestration. Thus, our research team explored the application of PSB and AMF in UA and investigated how they could benefit tomato production and soil health in UA.
Research approach
Our research aimed to answer the following questions.
- To discover how different combinations of AMF and PSB impact the growth and yield of tomatoes, soil nutrient cycling and plant nutrition, and the production of the soil-aggregating glomalin-related soil protein (GRSP).
- To compare microbial community compositions and metabolic functions using integrated omics studies on the soils of urban farms growing tomatoes under the treatment of AMF and PSB.
In 2022, we started to work with three urban organic farms located in New York City (NYC). Two farms, East New York Farm (ENYF) and Red Hook Farm (RH), were based in Brooklyn and New Root Farm (NR) was located in the Bronx. We used composts generated by each farm to grow tomatoes in a mixture of subsoil sand. We started to grow Bush Champion II Hybrid tomatoes (indeterminate variety) in 7-gallon-sized growing bags (Best Root Pouch, Maui Mike's Soft POTS) in three urban farms from 6/21 until 8/21 over the course of 10 weeks. The treatments were in five replicates for ENYF and six replicates for RH and NR.
- Control tomatoes treated with neither AMF nor PSB (Control tomatoes).
- Tomatoes treated with PSB (PSB tomatoes).
- Tomatoes treated with AMF (AMF tomatoes).
- Tomatoes treated with AMF and PSB (AMF+PSB tomatoes).
We employed commercial AMF and PSB products as our microbial sources to ensure that our research output would be more accessible to farmers if they were found to be effective. The commercial products were as follows:
- AMF product: DYNOMYCO® (DYNOMYCO), containing inoculums of Glomus intraradices and Glomus mosseae).
- PSB product: Mammoth P (Mammoth), comprising four taxa of PSB (Enterobacter cloacae, Citrobacter freundii, Pseudomonas putida, and Comamonas testosterone).
Succinct research conclusions
Our research has found that AMF and PSB had significant impacts on plant production and quality, soil bacterial communities, soil metagenome communities, and soil proteins and functions. Although more steps need to be taken, our current data generated consistently showed how AMF and PSB both shifted soil microbiome compositions and their co-abundance networks. Furthermore, changes in microbiome structure may explain how tomatoes treated by AMF and/or PSB result in lower root-to-shoot ratios (improved nutrient use efficiency), increased T-GRSP and acid phosphate activity, higher soil moisture, increased nutrient uptake, and different soil protein communities when compared to the control tomatoes.
Potential impact on the agricultural community
As far as agriculture is concerned, we believe that our research will provide valuable findings for both urban food producers and the scientific community. Firstly, our research is directed to support sustainable practice within urban agriculture, in that beneficial microbes can help with plant productivity and quality but also enhance microbial functions for nutrient uptake and cycling. Secondly, as we utilized commercial microbial products, our findings are reproducible in farms thanks to relying on products that can be purchased with ease. Additionally, little has been known about soil microbes and their functions within urban composts. Hence, our findings provide a good reference for upcoming microbiome studies based on urban agriculture.
Challenges and unexpected outcomes
In carrying out our research, our team was fortunate to have great collaborations with farmers who themselves had significant interests in the use of beneficial microbes. From the start, we have appreciated their support and dedication to our research. However, as our study took place on farms, dealing with the harsh weather conditions involved was challenging, unlike when growing tomatoes in a controlled growth chamber. Indeed, we began our work in the summer of 2022 when New York City experienced extreme heat weather events for a while. This made managing our plants problematic but, fortunately, we had a great research team helping each other to complete the tasks successfully.
Unexpected outcomes included an inability to count the number of tomatoes for a farm of our collaborating farms. Some people picked our tomatoes a few weeks before we harvested at a designated time. The farm is a community farm that welcomes a number of neighbors to join and volunteer in food production, so it is open to the public. Consequently, monitoring the farm site 24/7 was almost impossible, although we did ask all the people we met to kindly leave the tomatoes intact. Nevertheless, aside from this factor, we were able to preserve tomato plants and soils well. In comparison, other farms were fortunately protected by fences and gates, making it relatively easier to manage our tomatoes.
The overall aim of this research is to improve the physical and biological health of soils in urban tomato farms by increasing soil aggregation, nutrient cycling and availability, and microbial activity. The specific research objectives include:
1) To discover how different combinations of AMF and PSB impact the growth and yield of tomatoes, soil nutrient cycling and plant nutrition, and the production of the soil-aggregating glomalin-related soil protein (GRSP).
Reasoning:
This objective is designed to measure how AMF and PSB contribute to nutrient cycling in soil and plant nutrient uptake and growth. AMF are known to increase soil aggregation by producing a sticky and hydrophobic glue-like glycoprotein, GRSP, which can bind adjacent small-sized aggregates and enlarge them. Hypothetically, increased soil aggregation in soils becomes an energy source for the soil bacterial community to grow, thereby facilitating nutrient cycling and nutrient supply for plant uptake. The growth and yield of tomatoes can also be affected by increased nutrient availability in soils.
2) To compare microbial community compositions and metabolic functions using integrated omics studies on the soils of urban farms growing tomatoes under the treatment of AMF and PSB.
Reasoning:
This objective aims to elucidate how AMF and PSB treatments affect soil biochemical metabolisms and soil nutrient enrichment by examining soil microbiome changes and the protein expressions for nutrient cycling in urban agricultural soils. By integrating shotgun metagenomics with metaproteomics, we can identify how the inoculation of AMF and PSB creates a shift in soil microbial protein expressions in relation to soil nutrient metabolisms, and how this can relate to tomatoes’ nutrient uptake and growth.
The purpose of this project is to tackle chronic issues in urban agriculture such as poor soil structure and low nutrient availability using microbial resources to help produce higher economic returns to urban farmers. It is expected that more than 60% of the world’s population will inhabit urban areas by 2030. Urban agriculture holds great promise to feed such rising urban populations; in fact, urban agriculture is meeting from 2% to 150% of vegetables and fruit demands in urban areas. Urban agriculture is also critical for food security by providing cheaper and more nutritional foods to the urban poor.
However, many urban farmers face challenges in generating high yields and profits, because their soils lack good physical structure. They grow crops in raised beds using Technosol soils, which are comprised of mixtures of natural soil, compost, and soil-less media (e.g., perlite). These constructed raised beds often fail to develop sufficient soil aggregates where soil organic carbon (SOC) accumulates as aggregated structures that act as a reservoir for nutrients critical for plant uptake. Lacking SOC results in low soil fertility with low yields in urban agriculture, driving urban farmers frequently to add composts or fertilizers in an attempt to maximize their yields. Nonetheless, their attempts quite often lead to over-supply of nutrients in the form of highly mobile nitrogen (N) and phosphorus (P), which are subject to losses through waterways contributing to eutrophication.
In this proposal, we described a study to enhance soil nutrient cycling, soil aggregation, and plant growth and nutrition using two soil microorganisms: arbuscular mycorrhizal fungi (AMF) and polyphosphate solubilizing bacteria (PSB) to improve plant growth in urban soils that minimize environmental pollution. AMF can help develop soil structure by enhancing soil aggregation, while PSB could assist in soil P enrichment. Both soil parameters are important to maximize crop yields and farm net income, so AMF and PSB would promote the overall economic stability for urban farmers. Moreover, increasing nutrient cycling and soil structure would result in a reduction of nutrient loading to stormwater via runoff which will enhance environmental sustainability in urban agriculture. Our research is novel and will advance knowledge in the agricultural use of microbes in urban agriculture and the broader field crop systems.
Cooperators
- (Educator and Researcher)
Research
Our research was based on three urban farms including ENYF, RH, and NR in NYC. The research had four different microbial treatments—a control that does not contain either PSB or AMF; AMF only; PSB only; and AMF + PSB. We used commercial microbial inoculums because they are affordable and easily accessible to farmers. A commercial AM fungal product (DYNOMYCO®) was applied to the soils as inoculums of Glomus intraradices and Glomus mosseae. Another commercial product, Mammoth P, was used as a source of PSB which included Enterobacter cloacae, Citrobacter freundii, Pseudomonas putida, and Comamonas testosteroni. Each treatment was applied to a pot that contained different soil media that each farm had created. The compost was mixed with subsoil sand (70 compost : 30 sand) provided by NYC Clean Soil Bank managed by Office of Environmental Remediation. Subsoil sand is a soil that lacks organic matter, but we desired to improve its recyclability as a plant-growing substrate using beneficial microbes in UA where farmers can easily access urban construction sites and gain subsoil sand. We had to use the compost made by RH for growing tomatoes in both RH and ENYF because ENYF ran out of their compost at the outset of the experiment. However, as the farms were located far from each other, the sun conditions, growing weather, and surrounding environments were different. We were able to use the compost generated by NR for our research at NR. Bush Champion II Hybrid tomatoes were grown in 7-gallon-sized growing bags (Best Root Pouch, Maui Mike's Soft POTS) in three urban farms from 6/21 until 8/21 over the course of 10 weeks in 2022. The treatments were in 5 replicates for ENYF and 6 replicates for RH and NR.
Figure 1. Tomatoes at each farm during the 10-week growing period. Tomatoes were grown in 7-gallon growing bags and labelled with tags of different treatments. After 8 weeks, the tomatoes made heavy fruits, so we added wooden stakes next to several tomato pots and loosely tied each tomato to the stakes or the rope connecting the stakes in order to hold up the tomato and avoid fruit drop.
We initially prepared tomato seedlings from sterilized seeds using a fungicide and 70% ethanol. The tomato seedlings were inoculated with AMF and/or PSB depending on the treatment type and grown for 2 weeks in a growth chamber located at Cornell University. Then, we transported and transplanted the seedlings in growing bags filled with the respective farm compost. We added PSB in 1.5 gallons of water twice a week, following the optimal product-to-water ratio suggested by the company’s manual. Meanwhile, the Control tomatoes and the AMF tomatoes received only 1.5 gallons of water to which no PSB had been added. Figure 1 shows the chronological development of the tomatoes over the 10-week growing period and the layout of the tomato pot experiment at each farm site.
Figure 2. Research activities to address the aforementioned research questions.
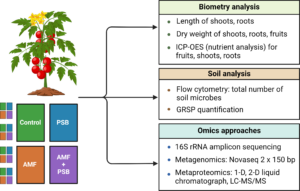
Figure 2 shows a list of the different experiments that were performed. Our research goal was to assess how external applications of AMF and PSB affect tomato growth and nutrition, soil health, soil microbial communities (microbiome), and soil protein communities (proteome). Thus, biometry analysis, including root and shoot length for each tomato, was performed on-site before the plants were harvested. The plants were dried for dry weight analysis and nutrient analysis using ICP-OES. Soil samples were kept at 4℃ to fix soil microbial cells and measure the absolute abundance analysis via flow cytometry as well as for the quantification of glomalin-related soil protein (GRSP). The soil samples that were used for the soil acid phosphatase (AP) activity analysis and omics work, including amplicon sequencing, metagenomics, and metaproteomics, were immediately frozen at -80℃ using dry ice upon harvest and kept frozen at -80℃ during transport and storage before analysis.
Objective 1: To discover how different combinations of AMF and PSB impact the growth and yield of tomatoes, soil nutrient cycling and plant nutrition, and the production of GRSP.
In order to achieve this objective, we planned to investigate multifaceted aspects of plant growth and soil health. This aspect of the research aimed to address our main question: whether a combination of AMF and PSB may produce the largest amount of GRSP for soil aggregation, enhanced nutrient cycling in soils, and plant nutrient uptake compared to individual treatments of AMF, PSB, or neither.
1) GRSP extraction and UV spectroscopy
The extraction of total GRSP (T-GRSP) was performed using repeated extraction from 1 g of ground soils kept at 4℃ with 8 ml of 50 mM citric acid, pH 8, at 121℃ for 1 h. At each autoclaving step, the supernatant was extracted and retained through centrifugation at 3,000 rpm for 10 min. The autoclaving and centrifugation steps were continued until the color of the extract turned yellowish, which signaled that all the GRSP had been extracted from the soil. The extracts of each cycle were pooled and centrifuged at 9,000 rpm for 10 min to remove all the soil particles. The extracts from each replicate were then analyzed using a Bradford assay with bovine serum albumin as standard on a 96-plate reader.
2) Plant tissue analysis and biometry measurements
Plant samples including roots, shoots, and fruits were oven-dried at 50℃ and ground and then prepared for the element analysis using ICP-OES. Biometry data of tomatoes, such as root and shoot height, shoot dry weight, and fruit dry biomass, were measured and compared in their total replications in all different treatments.
Objective 2: To compare microbial community compositions and metabolic functions using integrated omics studies on the soils of urban farms growing tomatoes under the treatment of AMF and PSB.
Little is known about bacterial communities inhabiting urban gardening soils in relation with the external usage of AMF and PSB. Our research aimed to discover the identity and genetic and biological functions of differently associated microbes with AMF and PSB that inhabited the soils of tomatoes in urban agriculture.
1) Flow cytometry for quantifying absolute abundance of soil microorganisms
Flow cytometry (FCM) was used to compare the absolute abundances of soil living microbes in different treatments. For FCM analysis, soil samples were suspended in phosphate-buffered saline (PBS) according to established protocols for soils. Total soil microbes were quantified in all conditions using flow cytometry using a dual staining method (using SYBR Green II and Tc). The quantification of total cells was performed using a BD LSR II available in the Institute of Biotechnology at Cornell University. Excitation at 488 nm generated forward scatter and side scatter of each cell, which was used to characterize different phenotypes of soil microbes and calculate absolute abundance.
2) 16S rRNA sequencing
DNeasy PowerSoil Pro Kits (Cat#: 47014; Qiagen, Carlsbad, CA) were used to extract the soil DNA from 0.25 g of the preserved soil samples at -80℃ following the manufacturer’s instructions. The 16S rRNA V4 hypervariable region was amplified by the universal primer set 515F (5’- GTGYCAGCMGCCGCGGTAA -3’) and 806R (5’- GGACTACNVGGGTWTCTAAT -3’), resulting in an amplicon size of ~253 bp in length. PCR reactions consisted of 10 μl of 2 x AccuStart II PCR ToughMix (QuantaBio, Beverly, MA, United States), 1 μl of 10 μM forward and reverse primers, 6 μl of DNase-free PCR Water (MoBio, Carlsbad, CA, United States), and 2 μl template DNA. The PCR thermocycling conditions were as follows: 94°C for 2 min; 25 cycles at 94°C for 20 s, at 55°C for 20 s, and at 72°C for 30 s; with a final elongation at 72°C for 5 min. Subsequence library preparation using unique two-barcode index combinations (Nextera, Illumina) followed by normalization was prepared on clear 96-well plates. The pooled libraries were paired-end sequenced (2 x 250 bp) using the MiSeq v2 500 bp kit (Illumina, San Diego, CA, USA) at the Cornell Genomics Facility (Ithaca, NY, USA).
Raw sequence assemblies and downstream analysis were performed using Qiime2 v.2023.2 (https://qiime2.org). After import, the amplicon sequence variants (ASVs) were demultiplexed and processed using Qiime DADA2 denoise-paired to trim reads, correct errors, and merge read pairs. Uchime was used to correct for amplicon sequencing errors and identify and remove chimeric reads, after which chimeric sequences were removed using an adapted Bayesian method. The 16s taxonomy assignment was performed by a Naive-Bayes classifier trained on the Greengenes2 2022.10 database using a q2-feature-classifier plugin. The resultant ASV dataset was imported into the R v4.1.2 using the Phyloseq package. The ASV abundance tables for each sample were then output into tab-delimited files. The ASV data were imported into R v4.1.2 and processed to perform multivariate statistics and diversity analyses.
3) Shotgun metagenomics
We performed shotgun metagenomic sequencing (i) to identify signals of specificity in the taxonomic and functional composition of the soil microbiota and (ii) to generate a reference library for the metaproteomic analysis. We generated metagenome-assembled genomes (MAGs) to assess the functional potential of the soil microbiota and to generate a protein sequence database for comparative metaproteomics across the two different treatments (the Control tomatoes and the AMF+PSB tomatoes from using methods that Dr. Manuel Kleiner (our collaborator) has successfully employed previously34.
In order to study functional dynamics, we required the full genome sequences of soil microorganisms. We will sequence these eight samples to a total read depth of 150bp paired-end reads (one full Illumina NovaSeq S4 flow cell to sequence these samples deeply enough for assembly and binning of MAGs. At all farms, our metagenomic sequencing was performed for the Control tomatoes and the AMF+PSB tomatoes in triplicates for the shallow sequencing. One pooled sample from the triplicates was deep-sequenced to aid with an in-depth understanding of the genomic potential and generate 255 million reads per sample. Thus, each site and treatment came to have 4 replicates for metagenomic sequencing.
We assessed read quality with FastQC and trimmed them using BBdduk to remove adapter sequences and low-quality regions. We assembled reads from individual or combined samples, comparing the output of the three metagenomic assemblers, MEGAHIT and metaSPAdes. We chose co-assemblies created by Megahit after comparing the quality of coasssembles of different assembly tools using Quast. We determined read coverage profiles for all contigs >1 KB across all metagenomic samples using BBmap and used MetaBAT to generate MAGs. We assessed the completeness and contamination of our MAGs using CheckM. For the purpose of generating a reference database for metaproteomic analysis, we retained all bins with <10% contamination, and also included unbinned scaffolds, provided they contain a reliable phylogenetic marker gene.
4) Metaproteomics
We prepared protein extracts from the soils of AMF+PSB and control groups, followed by peptide preparation for liquid chromatography (LC) mass spectrometry (MS)/MS analyses following the filter-aided sample preparation protocol. We used one dimensional (1D) or two dimensional (2D) LC and analysis of eluting peptides in a QExactive HF mass spectrometer as Dr. Manuel Kleiner has done previously. Mass spectrometric raw data were processed for protein identification using the ProteomeDiscoverer software (Thermo Scientific).
Statistical analysis
Statistical analysis included the Wilcoxon signed-rank test with Holm–Bonferroni correction for multiple comparisons (P < 0.05) to identify statistical differences between different combinations of PSB and AMF. Data were visualized via ordinations such as non-metric multidimensional scaling (NMDS) and principal component analysis (PCA) principal, network analysis, differential abundance analysis, and volcano plots to show variations in the bacterial metagenome and soil proteome. Heatmaps were also created to visualize and compare protein expression at taxonomic and functional levels between the treatments.
- Plant growth and soil health
Table 1. The table summarizing the biometric information of the plant and soil properties, including T-GRSP, soil AP activity, and soil moisture, of each treatment in NR, RH, and ENYF. NR and RH had six replicates, while ENYF had five replicates. Statistical significance was identified via the comparison of treatments within each farm group using the Wilcoxon signed-rank test with Holm–Bonferroni correction for multiple comparisons (P < 0.05). The data for the number of fruits were not available for RH because of unwanted fruit picking nearing harvest at 10 weeks. The average fruit dry mass of RH was based on the remaining fruits on each tomato until harvest.
|
Farm |
Treatment |
Shoot length (cm) |
Root length (cm) |
Root dry mass (g) |
Shoot dry mass (g) |
Root-to-shoot ratio |
Fruit dry mass (g) |
Number of fruits |
T-GRSP (mg/g) |
Acid Phosphatase activity (nmol/g hr) |
Soil moisture (%) |
|
NR |
Control |
57 a |
47.17 a |
3.06 a |
21.98 a |
0.16 a |
5.91 a |
4.17 a |
1.48 a |
329.19 a |
16 a |
|
PSB |
52.67 a |
52 a |
2.88 a |
27.02 a |
0.11 a |
6.32 a |
4.83 a |
1.76 a |
433.26 bc |
21 bc |
|
|
AMF |
59.17 a |
48.67 a |
4.04 a |
22.3 a |
0.17 a |
6.99 a |
5.17 a |
2.18 a |
383.53 b |
24 b |
|
|
AMF+PSB |
59.67 a |
40 a |
3.61 a |
24.15 a |
0.15 a |
4.89 a |
4.83 a |
2.12 a |
460.52 c |
20 c |
|
|
RH |
Control |
66.67 a |
64.5 a |
4.32 a |
36.75 a |
0.13 a |
4.99 a |
NA |
1.46 a |
138.25 a |
16 a |
|
PSB |
73.83 ab |
62.77 ab |
7.17 ab |
39.83 a |
0.2 a |
6.29 a |
NA |
1.56 ab |
171.61 b |
16 b |
|
|
AMF |
83.5 b |
75.83 b |
5.96 a |
52.9 a |
0.11 a |
7.21 a |
NA |
2.04 b |
196.2 c |
23 c |
|
|
AMF+PSB |
87.5 ab |
71.12 ab |
7.82 b |
49.63 a |
0.17 a |
6.81 a |
NA |
2.43 c |
200.48 c |
17 c |
|
|
ENYF |
Control |
64.6 a |
64.8 a |
9.42 b |
47.68 a |
0.2 a |
5.87 a |
4.6 a |
1.71 a |
108.89 a |
10 a |
|
PSB |
66.6 a |
59.08 a |
7.94 ab |
53.36 a |
0.16 ab |
5.26 a |
7.6 a |
1.61 a |
120.67 ab |
17 ab |
|
|
AMF |
64.2 a |
72.6 a |
7.34 ab |
45.92 a |
0.16 ab |
5.22 a |
6.2 a |
2.07 a |
134.57 b |
18 b |
|
|
AMF+PSB |
69.2 a |
49.78 a |
5.61 a |
44.24 a |
0.13 b |
3.69 a |
10.8 a |
2.07 a |
166.51 c |
18 c |
Table 1 presents a summary of the tomatoes’ growth parameters, including shoot length, root length, root dry mass, shoot dry mass, root-to-shoot ratio, fruit dry mass, and number of fruits in addition to soil characteristics such as T-GRSP, AP activity, and soil moisture. We did not detect consistent trends regarding the impacts of AMF and PSB on the tomatoes’ physiological growth and development. In RH, adding AMF significantly increased the lengths of the roots and the shoots in the AMF tomatoes and the AMF+PSB tomatoes compared to the Control tomatoes. In contrast, the AMF+PSB tomatoes in ENYF had smaller root dry mass compared to the Control tomatoes, while the root-to-shoot ratio of the AMF+PSB tomatoes was significantly smaller than that of the Control tomatoes.
The production of GRSP increased when tomatoes received AMF or AMF+PSB in RH. Even though statistically insignificant, both NR and ENYF showed an increasing trend regarding the production of T-GRSP in soils as compared to the non-AMF tomatoes, i.e. the Control tomatoes and the PSB tomatoes. GRSP is a soil aggregating protein and AMF may be involved in its production in soils. We observed that all tomatoes treated by AMF had increased amounts of GRSP in the soils at all farms. This may indicate that AMF can help soil aggregation and maintain the physical structure of urban compost soils.
The activities of AP were higher in the tomatoes inoculated with PSB and AMF at all farms. AP is synthesized by soil microbes and plants and helps release more P in an available form for plants and enhances P availability. Our findings suggest that AMF and PSB contributed to AP activities in the rhizosphere of the tomatoes, which may have led to better P usage by the tomatoes.
Soil moisture was also higher in the tomatoes treated by AMF and PSB compared to the Control tomatoes on all farms. Thus, tomatoes treated with AMF and PSB can maintain water for longer in the urban compost type of soils, which are otherwise often susceptible to drying due to their poor physical structure.
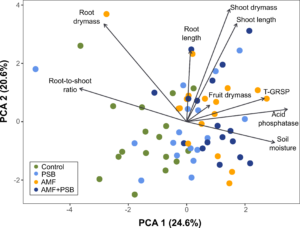
Figure 3. Biplot of the PCA of the plant and soil metadata (Table 1). Each value within each dataset was scaled and centered around the mean with a unit variance. Scaling was performed in order to remove the variations of different farms and enable a larger-scaled comparison of the different microbial treatments across the different farms. The site scores are colored depending on their treatment type. The direction of each vector (arrow) is related to the direction of increasing correlation with site scores. The closer the site point is to an arrow, the higher the values of that variable they have.
We observed that there was no statistical significance for some data in Table 1 using the Wilcoxon signed-rank test given the confidence level. However, as each site had low replicate numbers (low sample size), we wanted to expand our statistical power into a metadata scale by scaling each farm data set centered on their mean value and combining all farm data for statistical testing.
Figure 3 shows the patterns of the treatment variations determined by various plant and soil characteristics identified via PCA based on the metadata. The PCA ordination revealed that plant and soil data can explain 45.2% of the total variance of the dataset. Each arrow represents the correlation of each parameter with the variations explained by PCA. It means that the longer the vectors, the more influential they are. We found that most of the samples from the Control tomatoes (green dots) were located in opposite directions in terms of root length, shoot dry mass and length, T-GRSP, and AP production. This indicates that although our dataset did not identify significant differences among the treatments on a given farm in low replicates, there was a trend in that the tomatoes treated with AMF and/or PSB outperformed the Control tomatoes in terms of plant growth and development after the data had been standardized and compared on a multivariate scale.
Many tomatoes treated by AMF and/or PSB were closely correlated to high soil moisture compared to the Control tomatoes. It may support that the inoculation of AMF and PSB could improve soil moisture retention and drought resistance of plants in UA.
The control tomatoes were more closely associated with longer root-to-shoot ratios, while those treated by AMF and/or PSB tended to have lower root-to-shoot ratios. This may indicate that tomatoes inoculated with AMF and/or PSB may use nutrients more efficiently so do not need more investment in root development – instead, allocating more energy resources to shoot development.
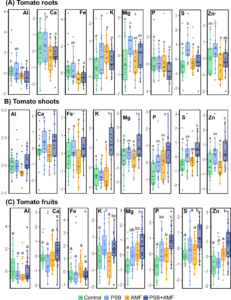
Figure 4. (click on image to open in a new tab.) Nutrient compositions of different plant tissues identified by ICP-OES. All nutrient data from the same farm were scaled before statistical difference was identified using the Wilcoxon signed-rank test at Holm–Bonferroni modified P < 0.05. The nutrient contents of RH were not included in the analysis yet.
We performed ICP-OES to see the distribution of plant nutrients such as aluminum (Al), calcium (Ca), iron (Fe), potassium (K), magnesium (Mg), phosphorus (P), sulfur (S), and zinc (Zn) in the different parts of the plants, including roots, shoots, and fruits (Figure 4). We did not detect significant differences between the tomato treatments for the nutrients of the roots, except for Al, Fe, S, and Zn. However, K, Mg, P, S, and Zn were all higher in the shoots and fruits of the tomatoes inoculated with both AMF and PSB compared to the Control tomatoes. The relatively weak differences in the root samples may relate to the translocation of important nutrients from the roots to the shoots as the fruits matured. We discovered that the AMF+PSB tomatoes were more efficient in using important plant nutrients for fruit production than the Control tomatoes.
2. Changes in soil microbiome structures
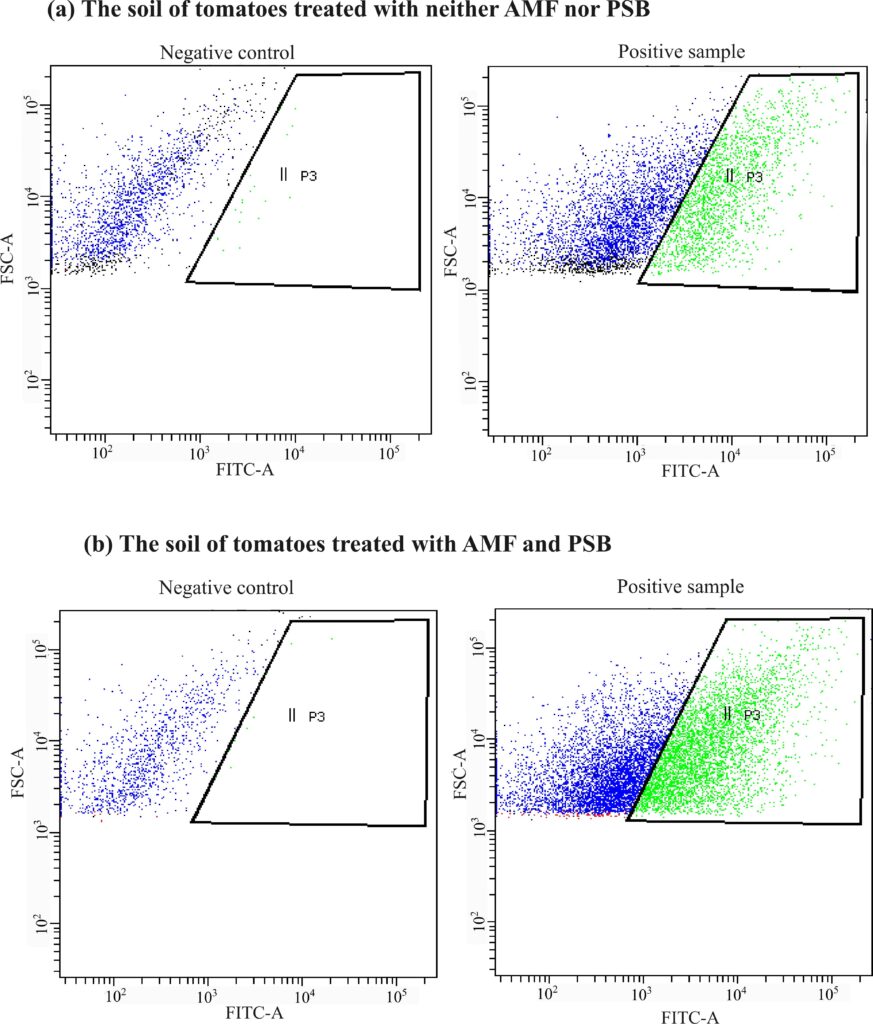
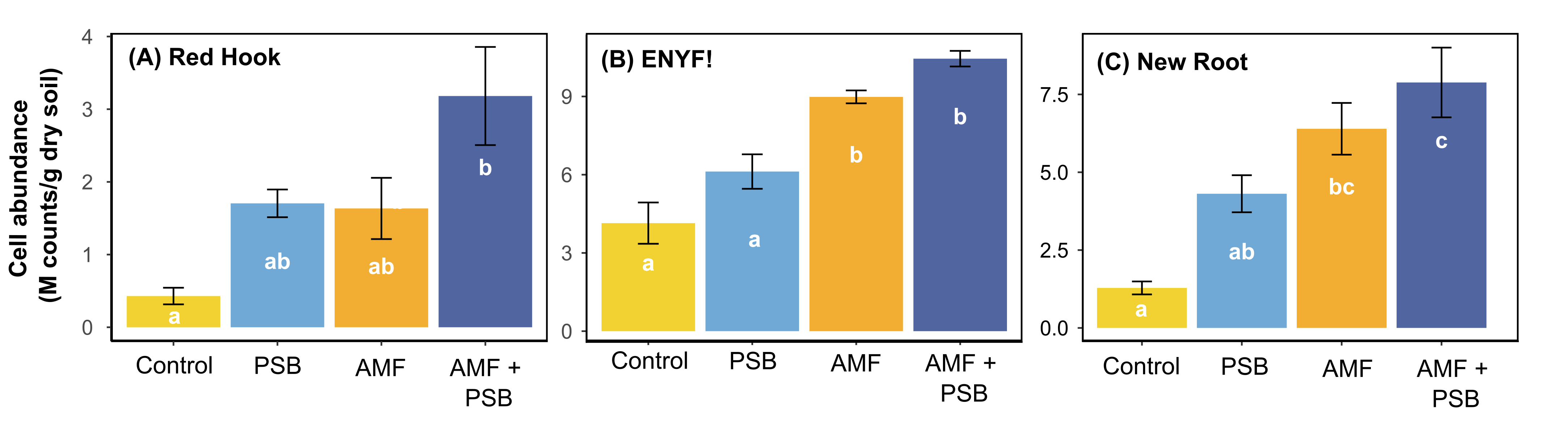
Figure 5. Flow cytometry results showing the number of cells detected in the root microbiome in the soil of tomatoes. The negative controls show the population of negative cells that are unstained by SRBR green I and that could be gated out in the assay for positive samples. (a) The tomatoes treated with neither AMF nor PSB had a relatively lower abundance of cells than (b) the tomatoes treated with both AMF and PSB.
Figure 6. The total number of millions (M) of living microbial cells of the soil treatment at (A) Red Hook Farm, (B) ENYF, and (C) New Roots using flow cytometry: soil microbial biomass was stained with SYBR Green and represented as a green dot.
We observed that the absolute abundance of soil microbes significantly increased in the AMF and PSB tomatoes compared to the Control tomatoes on all farms (Figures 5 and 6). At all farms, the AMF+PSB tomatoes had statistically higher numbers of soil microbial cells. Thus, the data explained that adding external microbial sources to tomatoes could lead to increasing the total number of soil microbes (absolute abundance of soil microbiome). Soil microbes play essential roles in improving nutrient cycling, and thus increased numbers of soil microbes in the AMF+PSB tomatoes can benefit the overall microbial activities in the soil.
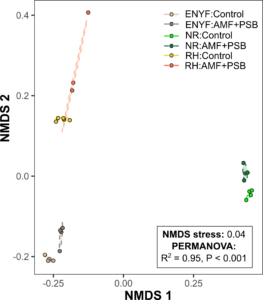
 Figure 7. The NMDS ordination of soil bacterial communities of different farms composts. Pairwise PERMANOVA analysis was employed to detect statistical differences among the treatments, whereby the different numbers of asterisks indicate significance levels: * < 0.05; ** < 0.01. Site scores are in five different colors. The ‘Original’ site is the soil microbiomes of each compost at each farm site collected before the experiments, and thus represents the soil bacterial communities of the original compost.
Figure 7. The NMDS ordination of soil bacterial communities of different farms composts. Pairwise PERMANOVA analysis was employed to detect statistical differences among the treatments, whereby the different numbers of asterisks indicate significance levels: * < 0.05; ** < 0.01. Site scores are in five different colors. The ‘Original’ site is the soil microbiomes of each compost at each farm site collected before the experiments, and thus represents the soil bacterial communities of the original compost.
We identified that growing tomatoes and adding AMF and/or PSB caused significant changes in soil bacterial community structure (Figure 7). At all farms, the soil microbial compositions shifted far from the original microbial population. In addition, tomatoes treated with AMF and/or PSB had microbial community structures that differed from each other and from that of the Control tomatoes. This indicates that AMF and PSB both actively interacted with soil microbiota in unique ways.
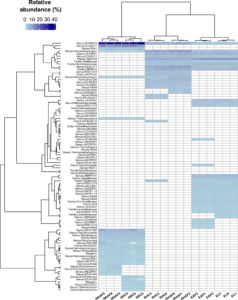
Figure 8. (click on image to open in a new tab.) The phylogenetic tree of microbes accounting for more than 0.5% relative abundance. The relative abundance of each species at each treatment site was presented on a gray-scale heatmap. The maximum relative abundance of each species was described in a horizontal bar plot and colored by the site at which they were most abundant. Taxonomy was assigned to each species at its lowest rank.
The phylogenetic tree was created based on soil microbes that accounted for more than 0.5% relative abundance and occurred in more than 80% of all replicates of the same treatment (Figure 8). The heatmap of relative abundance roughly shows that ENYF and RH shared quite a lot of microbial species because the starting soil composts were basically the same. However, we identified that both ENYF and RH developed microbial communities that were more specific to each site because of the site differences. NR had a microbial community that was distinct from those of EBYF and RH. Some species were shared in all soil types, including the day 0 original soils. They were Class:Bacilli, Genus:Geobacillus, Genus:Planifilum, Genus:ZCTH02-B6, Order:Thermomicrobiales, Family:Micromonosporaceae, Genus:SZUA-442, and Genus:RSA9. These species may represent a group of soil bacteria that are ubiquitously abundant in urban soil composts.
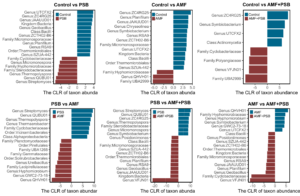
Figure 9. Differential abundance analysis of soil bacteria. The bar colors represent where the species were more abundant. The analysis was based on microbes that occurred in more than 80% of the total sample replicates of the same group of all farms. The comparison was based on the centered log ratio (CLR) of the abundance of each species. We only showed the species that had an absolute effect size of difference higher than 2 and whose abundances were found to be significantly different between groups based on the Wilcoxon test with Benjamini-Hochberg correction, P < 0.05.
We performed differential abundance analysis using Aldex2 and discovered that many microbes were differentially abundant across different microbial treatments (Figure 9). Adding AMF and/or PSB to tomatoes caused significant differences in the abundance of microbes that interacted with plants, AMF, and PSB compared to the Control tomatoes. Presumably, those species that were differentially abundant may have contributed to the differences between the Control tomatoes and the tomatoes treated by AMF and/or PSB.
Furthermore, significant differences were found between the PSB and AMF tomatoes, the PSB and AMF+PSB tomatoes, and also the AMF and AMF+PSB tomatoes. There were significant differences between the PSB tomatoes and the AMF tomatoes, between the PSB tomatoes and the AMF+PSB tomatoes, and between the AMF tomatoes and the AMF+PSB tomatoes. This confirms that AMF and PSB have their own ways of communicating with other soil bacteria. The dual inoculation created a distinctive microbial structure different from a single inoculation of both, showing the presence of interactions between AMF and PSB. We already observed enhanced food quality and higher nutrient uptake in the fruits of the AMF+PSB tomatoes compared to the Control tomatoes. Thus, not only did the dual inoculation influence the soil microbial community structure but also their interaction with other soil microbes might have led to increased nutrient availability and nutrient uptake by the plants.
Figure 10. (Click on image to open in a new tab.) Network analysis based on bacterial communities of different tomato treatments. The number of nodes is the number of microbial interactions associated with each species – described by a differing circle size. The SparCC correlation coefficient represents the strength of those interactions; the higher the coefficient, the stronger its co-abundance with connected species. The top 20 nodes that had the highest microbial association were selected for each bacterial co-abundance network. All interactions were identified at P <0.05.
We aimed to find out how different microbial treatments affected the microbial interactions in the soils. To achieve this, we performed SparCC network analysis and identified significant microbial interactions of the top 20 species – in terms of the number of interactions with other species (Figure 10). We discovered that microbial interactions significantly changed when tomatoes were inoculated by AMF and/or PSB. Some species were actively involved in the co-abundance networks, for example, Genus:Bacillus, which appeared to be abundant with others in the top 20 networks of the Control, PSB, and AMF tomatoes alike. However, in the network of the AMF+PSB tomatoes, this genus was not included in the top 20 microbes. Thus, not only did AMF and PSB affect the microbial structure but they also changed the microbial interactions, which led to an investigation of how soil microbial functions may change accordingly.
3. Changes in soil functions
Figure 11. NMDS ordination plot for the metagenomes of the Control tomatoes and the AMF+PSB tomatoes. Each treatment is composed of 3 shallowed sequenced samples and 1 pooled sample.
We quality-checked the raw metagenomes and then performed phylogenetic analysis using PhyloFlash. Using the PERMANOVA test, we then identified significant differences in terms of metagenome composition between the Control tomatoes and the AMF+PSB tomatoes (Figure 11). The soil metagenome used for the heatmap includes not only bacteria but also fungi and eukaryotes. Thus, our metagenomes show that treating AMF and PSB tomatoes has the potential to change the collective microbiome compositions, including for both bacteria and fungi.
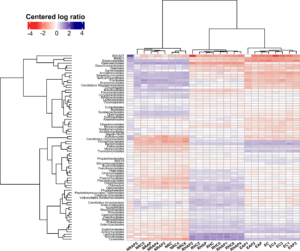
Figure 12. Heatmap of the relative abundance of species across different treatments, transformed in centered log ratio. The relative abundance of species was generated by PhyloFlash. Abbreviations include NRAP: AMF+PSB tomatoes of New Root; NRC: Control, New Root; RHAP: AMF+PSB, Red Hook; RHC: Control, Red Hook; EAP: AMF+PSB, East New York Farm; and EC: Control, East New York Farm.
The heatmap shows that all samples of the same farm were clustered together in which the samples of the Control tomatoes and the samples of the AMF+PSB tomatoes of the same farm were clustered separately (Figure 12). At all farms, we found that the soil metagenome community structures of the Control tomatoes were significantly different from those of the AMF+PSB tomatoes.
Table 2 summarizes the number of MAGs identified at each farm. Overall, we found 1,257 MAGs in ENYF, 1,492 MAGs in NR, and 1,635 MAGs in RH, where many bins were classified as medium-to-high quality. We plan to submit our bins to public database, because there have not been many studies on soil microbiome of urban gardening soils. Our data will be a good reference for future studies on UA.
Table 2. Quality assessment of bins using CheckM. The number of bins by different quality criteria discovered in different farms.
|
Bin quality |
Criteria |
ENYF |
NR |
RH |
|
Medium to High |
Completion >50, Contamination < 10 |
677 |
790 |
731 |
|
Low to Medium |
Completion > 30, Contamination < 5 |
82 |
170 |
194 |
|
Bad bins |
Completion < 30, Contamination > 5 |
356 |
248 |
385 |
|
Unbinned |
The rest |
142 |
284 |
325 |
|
Total |
1257 |
1492 |
1635 |
|
The following heatmap is based on the relative abundance of MAGs (Figure 13). Similar to the heatmap (Figure 12), each treatment has a similar distribution of MAGs and the samples that belonged to the same farm (both samples from the Control tomatoes and the AMF+PSB tomatoes) were clustered closely. However, some MAGs were differentially abundant between the Control and the AMF+PSB tomatoes. We believe that the differentially abundant microbial species between the treatments may determine different microbiome functions. Indeed, we had already observed differences between treatments for physiological aspects (Figure 3), as well as for nutrient uptake and fruit quality (Figure 4). Microbial functions were closely related to the collective functions of soil protein communities. Thus, we aimed to study soil proteins using microbial functions as a metagenomic reference.
Figure 13. Heatmap of the relative abundance of metagenome-assembled genomes (MAGs) across different treatments transformed in centered log ratio. Abbreviations include NRAP: AMF+PSB tomatoes of New Root; NRC: Control, New Root; RHAP: AMF+PSB, Red Hook; RHC: Control, Red Hook; EAP: AMF+PSB, East New York Farm; and EC: Control, East New York Farm. GTDBTK was used to assign bins to microbial taxonomic groups at their lowest rank.
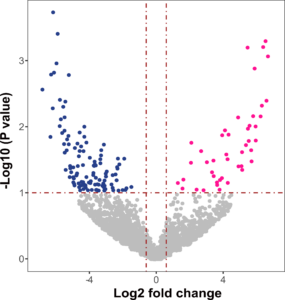
Figure 14. Differentially abundant proteins in the rhizosphere soil of the control tomatoes (pink) and the AMF+PSB tomatoes (blue) at ENYF are highlighted in color (Log2 fold change > 0.6 at P < 0.1).
Currently, we are in the process of identifying functional proteins shown to be differentially abundant across treatments. Figure 14 is one of the outputs based on the ENYF soil proteins. The coassembled contigs were used to identify the proteins extracted from the soils. In doing so, we have found quite a lot of proteins to be differentially abundant between the Control tomatoes and the AMF+PSB tomatoes. Our research provides evidence proving coherently that AMF and PSB cause some physiological differences in tomatoes, soil bacterial communities, soil metagenomic communities, and soil protein functions. Our next step is to elucidate what proteins are differentially abundant and which microbes are involved in producing those proteins and their functions in each tomato treatment.
Our research has discovered that AMF and PSB tomatoes were more associated with lower root-to-shoot ratios, higher production of T-GRSP, high AP activity, improved soil moisture, enhanced nutrient uptake, and better fruit quality. They affected soil bacterial communities differently, with lots of bacteria proven as differentially abundant depending on how AMF and PSB were treated. Because of this, the soil bacterial co-abundance networks formed differently, which may cause interactions that change soil microbial functions. Our multi-Omics work has identified a total number of 4,384 MAGs (each represents a full genome sequence of a single microorganism) from three farms. The MAGS distribution produced consistent results with the 16S bacterial communities, showing the Control tomatoes to have different microbial compositions from the AMF+PSB tomatoes. Our metaproteomic work is still ongoing but has already identified differentially abundant proteins between the Control tomatoes and the AMF+PSB tomatoes. Our work is still ongoing to identify the protein identities and assess how they are related to different microbiome structures, as well as plant growth and development performance. More details about it will be explained in a published paper soon.
We believe that our findings will be important for future urban agricultural work. Our work can act as a reference for future microbiome studies based on urban agricultural compost soils. In addition, we have discovered that AMF and PSB can both improve plant productivity, particularly in terms of fruit and soil quality. Urban compost soils are known to lack the microbial functions required for recycling nutrients and facilitating nutrient uptake for a plant. However, our study shows that adding external microbial sources such as AMF and PSB can help to improve plant development and soil health by showing how they were able to transform soil microbial composition and functions. Furthermore, our study encourages more extensive use of beneficial microbes in urban agriculture to support the sustainable production of healthy food in urban areas.
Education & Outreach Activities and Participation Summary
Participation Summary:
The outreach plan for this research had three components: 1) working closely with farm staff and youth interns, supporting youth in sharing research results and applications with local NYC farmers and networks, 2) sharing findings and applications with regional farmer and Extension networks, and 3) sharing detailed analytical results with national and international academic audiences.
This research will builds upon and extends many of the goals the large Cornell Agricultural Soils of the City project (https://blogs.cornell.edu/nyurbansoils/) to better understand and support the sustainability and productivity of urban agriculture. Specifically, we worked with three NYC farms that have been part of this project: East New York Farms! (ENYF), Red Hook Farms, and New Root Farms. Youth and farm staff were consulted in the planning of the research and helped establish plots, tended to crops, and assisted with sampling and harvest.
Ongoing outreach beyond the Northeast SARE project scope will include meetings with collaborating farmers and youth about research data and co-creation of presentations for Northeast Organic Farming Association and Cornell Cooperative Extension’s Ag-In Service Conference. Regional urban agriculture endeavors are also expanding, and this research will be shared with these audiences at conferences such as the 2024 Urban Food Systems Symposium. We anticipate that urban farmers and organizations in other cities will not only be interested in the impacts of AMF and PSB, but will also be interested in enhancing soil microbial activities and plant quality. We will write a series of relevant Extension documents that can be shared with these networks.
Finally, the results of this research will be submitted to peer-reviewed journals such as Science Advances, as our research used an innovative multi-omic approach to reveal soil microbiome and protein functions in UA. The results presented in this manuscript will be addressed towards academic researchers, who we anticipate will be interested in the novel approaches that we use both in terms of investigating PSB and AMF in agriculture and in the range of analytical methodologies that we employ. These results will also be shared at academic conferences including the annual meeting of the Soil Science Society of America as well as the international Soils of Urban Industrial, Traffic, and Mining Areas conference.
Project Outcomes
Our previous microbiome survey of nine urban farms in NYC reported that many urban farms fail to establish good soil structures containing balanced nutrient reservoirs. This could represent a critical obstacle for creating high-yielding food production within UA, resulting in a heavy dependence on chemical fertilizers in urban farms. Our research has discovered that inoculation with AMF and PSB can improve tomato growth, soil health, and change soil microbiomes and their functions. Thus, our discovery evidences that adding external sources of microbes such as AMF and PSB can improve plant productivity and quality in UA. Farmers can easily obtain commercial microbial products from markets and try them to increase their farm productivity. Microbial fertilizers are not only harmless to the environment but also helpful in reducing the heavy reliance on expensive agrochemicals and promoting sustainable agriculture in UA.
Our research discovered that tomatoes inoculated with AMF and PSB were able to produce more GRSP and obtain plant nutrients from the soils. Thus, the tomatoes inoculated by AMF and PSB had more efficiency in terms of taking up nutrients and allocating them to shoots and fruits, offering higher major market value for horticultural production. Many nutrients in UA soils are often subject to leaching into the environment and the groundwater system, causing significant environmental issues such as eutrophication. Allowing a significant amount of nutrients, such as P, to leach into the municipal waste stream can put urban well-being and the health and safety of NYC residents at risk. Thus, using microbes can be beneficial as they reduce nutrient leaching through enhancing plants’ nutrient uptake as well as soil aggregation (caused by GRSP).
What we learned about sustainable agriculture from working with urban farmers
We have been working with good people, including excellent research groups who helped this research and great and passionate farmers who are interested in beneficial microorganisms. Farmers were greatly interested in our research and in improving sustainability via their farm practices. The research idea started from pure curiosity about how beneficial microbes can benefit tomato production. However, after several discourses with the collaborating farmers, we understood that they are passionate about my research and want to learn more about beneficial microbes for own their plant production. Thus, we learned that to truly improve sustainable practice, it is important for researchers to work closely with farmers. This involves frequent communication with them to understand their needs and build trust based on their passion for this project. Thanks to our wonderful research collaborators, we have gained substantial knowledge on how our research could be impactful and more effective in improving sustainable practices in farms. The collaborating farmers, including their youth, were involved in all the procedures of the growing, managing, and harvesting of our tomatoes. They were excited to try AMF and PSB on their plants in the coming 2024 growing season.
The future direction of career and research
The ultimate career goal of our research team is to improve plant production in a way that our nature and environment are more sustainably managed in the long term. Our research team is passionate about sustainable agriculture and believes that soil microbes will play important roles in horticultural production. The roles and functions of soil microbes need more research to fully utilize their unexplored potential for soil nutrient cycling and plant production. This research has built an important step toward accomplishing our goal of promoting the use of beneficial microbes for horticultural production in UA.