Final report for GNE22-279
Project Information
Since June 2023, all US livestock producers need a veterinarian-client-patient-relationship (VCPR) to be able to get a prescription to purchase any antimicrobial to treat and manage disease. The combination of limited livestock veterinarians and the lack of veterinary use by small ruminant producers creates urgency in identifying alternative strategies to combat disease. The utilization of estimated breeding values within the Katahdin breed has enabled sustainable improvement in resistance to parasites. Recent data indicates that selection for post-weaning worm egg count (PFEC) estimated breeding value (EBV) improves lamb survivability to weaning. Additionally, antibody response to vaccination was greater in Katahdin lambs with lower PFEC values. Taken together, these data provide evidence to indicate that Katahdin sheep with lower PFEC values are more immune competent. To test this hypothesis outside the context of parasitic infection we would need to model a bacterial infection, which can be achieved by using bacterial cell wall products like lipopolysaccharide (LPS). Thus, the objective of this project was to determine effects of PFEC genotype on immune response to LPS. To fully characterize this response, in vitro methodologies were utilized to evaluate transcriptome-wide differences between PFEC genotypes in response to LPS. This data was used to support implementation of selection strategies that incorporate PFEC and by doing so potentially reduce the need for and use of antimicrobials. Reducing disease, the need to treat disease, and preserving the efficacy of medically important antimicrobials is fundamental to the sustainability of small ruminant production.
The approach of this project can be clarified by the objectives below:
- To characterize differential immune response between LoPFEC and HiPFEC sheep to LPS stimulation in-vitro
- To determine transcriptome-wide gene expression differences between LoPFEC and HiPFEC-derived cells stimulated with LPS
Funds requested from this proposal were principally used to characterize the immune response and perform transcriptome-wide analysis in an effort to elucidate patterns in Katahdin sheep with divergent genotypes for PFEC EBV. This project was used as a starting point to allow for a comprehensive genetic analysis of the Katahdin breed in the context of bacterial infections. By using a transcriptome-wide approach, we gained new insights into the differences in gene regulation in sheep bred for PFEC, which had strong preliminary evidence to regulate immune function. This provided potential avenues of exploration into the immunoregulation of individuals during a disease state. Fundamentally, we aimed to substantiate the claim that selection for PFEC selects for improved immunocompetency outside of just parasitic infections. This work was presented in the context of improved selection for animal health to producer groups both regionally and nationwide. These findings are being analyzed for publication, with plans to submit by the end of the year.
The purpose of this project is to evaluate the effect of selection for parasite resistance on improved generalized immune responsiveness in sheep as a method to reduce the need for antimicrobial use. The use of antimicrobials (AM) in livestock production has been significantly reduced since implementing the veterinary feed directive, which made the use of AM in feed and water prescription based. To further reduce AM use by livestock producers, the FDA will regulate sale of over-the-counter AM to be by-prescription-only by June of 2023, in efforts to mitigate development of AM-resistance. Antimicrobial resistance is at the forefront of the FDA’s Center for Veterinary Medicine (CVM) challenge to national and worldwide public health (Patel et al., 2020). As part of its regulatory mission, CVM is responsible for ensuring the safety and effectiveness of animal drugs, including AM. The CVM has taken important steps to update the approved use conditions of medically important antimicrobials (i.e., antimicrobials important for treating human disease) to support their judicious use in food-producing animals (Patel et al., 2020). While most antibiotic use in livestock requires a prescription or a veterinary feed directive, individual decisions on administration are often made by farmers via guidelines provided by a veterinarian. Currently, surveillance data do not indicate dosages, or whether antibiotics were administered to prevent, control, or treat a disease (Patel et al., 2020). According to the National Resources Defense Council's (NRDC) analysis of the most recent data, some of the pitfalls surrounding antibiotic stewardship include animal husbandry practices on farms and the level of farm antibiotic-utilization rates (Wallinga and Kar, 2019). This indicates that livestock producers may inadvertently be misusing antibiotics and contributing to the emergence of resistant bacteria. Therefore, reducing AM use in animal agriculture is paramount to the sustainability of the livestock industry.
Antimicrobial stewardship at the producer level relies on implementation of non-AM disease control strategies. Currently, producers have limited options of non-antimicrobial strategies to utilize within their flock. The research proposed here intends to identify genetic underpinnings which account for differences in immune response in sheep bred to be parasite resistant. Parasite resistance is reported as an estimated breeding value (EBV) for post-weaning worm egg count (PFEC). Previous data indicate that Katahdin sheep with low PFEC (parasite resistant) have greater survivability to weaning. Since significant variability for PFEC exists within the Katahdin breed (-100 and +867) (NSIP Database, 2022), the opportunity for selection would allow producers to capitalize on genetic tools to implement quick progress. By investigating resistance mechanisms involved in differential PFEC genotypes, we will be better positioned to provide producers with an effective means to implement non-antimicrobial disease prevention within their flocks. Selection for LoPFEC will have sustainable outcomes on sheep production by improving animal health and reducing the need for AM use.
Cooperators
- (Educator)
Research
General Approach: Katahdin ewes were sourced from the Southwest Virginia Agricultural Research Extension Center (AREC) located in Glade Spring, Virginia. All individuals had NSIP data and index values that were used to ensure ewes are relatively similar in genetic merit beyond PFEC. To achieve this goal, lambs must have had a minimum Katahdin Index of +104. Once that threshold had been met, then 10 sheep of each genotype were randomly selected after they met the following conditions. Low PFEC group (LoPFEC) was required to have a PFEC EBV of less than -50, and high PFEC sheep (HiPFEC) was required to have a PFEC EBV of greater than +200. In both instances, accuracy of PFEC estimates must be equal to or greater than 75%. Once identified, these lambs were relocated to WVU Agronomy farm.
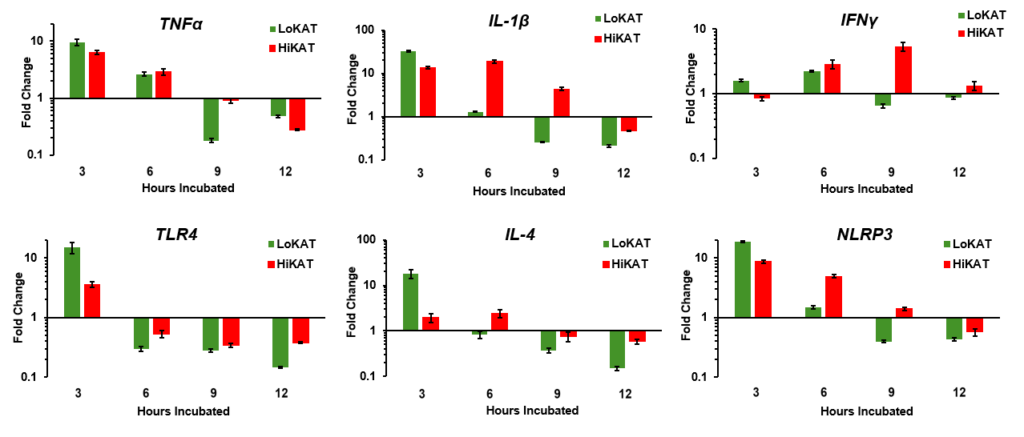
Objective 1: These sheep were then used to collect peripheral blood mononuclear cells (PBMCs) for in vitro lipopolysaccharide (LPS) stimulation. To determine immune cell response to LPS, peripheral blood mononuclear cells (PBMC) were isolated and quantified using a TC-20 automated cell counter (Bio-Rad, Hercules, CA). PBMC was diluted in RPMI-1640, supplemented with fetal bovine serum and antibiotic. PBMC were added to wells in triplicate by individual for both control and LPS treatments. Cells were treated with 100µg/ml of LPS derived from E. coli O111:B4 (L4391, Sigma Aldrich, St. Louis, MO). To determine differences in response to LPS activation, we measured typical cellular response to LPS stimulation. PBMC (1x105) collected from each animal were plated in triplicate and treated with LPS (100ug/ml). PBMC with and without stimulation were incubated for 3, 6, 9, and 12 hrs. At each time point cells were collected, and RNA extracted they were converted to cDNA for Real-time PCR gene expression analysis. Our previous in vivo work has shown that LoPFEC sheep had greater serum levels of Tumor Necrosis Factor alpha (TNFα) than HiPFEC sheep for 8 hrs after LPS administration. Gene expression of immune cytokines (TNFα, IL-1β, IFNγ, IL-4) and other immune mediators (TLR4 and NLRP3) over a time course allowed us to determine the time point where the greatest difference exists between LoPFEC and was able to provide critical information to accomplish project objectives. Data from this study was analyzed using proc mixed procedure of SAS 9.4 (Cary, NC) with time, treatment, and genotype as main effects. Interactions were analyzed using appropriate post hoc procedures and significance is accepted when P < 0.05.
Objective 2: Once this optimum timepoint was determined, PBMCs was once again be collected and stimulated (Control vs. LPS) for this amount of time and RNA was isolated from these cells. For this analysis, 5 sheep from each genotype were be randomly selected so that a total of 20 RNA-seq analyses were to be performed (10 LoPFEC; 5 control and 5 LPS-treated and 10 HiPFEC; 5 control and 5 LPS-treated). This RNA was submitted to the WVU Genomics Core for cDNA library build and RNA sequencing. Good quality reads were then aligned against the ovine reference genome (Oar_v4.0) using STAR, and unique reads were quantified. Alignment mapping analysis was conducted to determine the differences between the reference genome and the sequences provided by our experiment. This data was normalized, and statistical analyses performed using DESeq2 (negative binomial distribution, outlier detection, and correction). The Benjamini-Hochberg procedure was performed to obtain an adjusted P value (Benjamini-Hochberg adjusted P value) with a significance cut-off of P = 0.05. Following this we compared gene upregulation in the LoPFEC versus HiPFEC resulting from LPS stimulation. This gave us the potential to evaluate the entire transcriptome and discover genes previously not known to be involved within the phenotypic differences we observe through divergent PFEC selection.
TNFα (Tumor Necrosis Factor Alpha) is an important pro-inflammatory cytokine that plays a key role in the body's immune response and inflammation. It is intriguing that TNFα levels are elevated at 3 hours in PBMCs (peripheral blood mononuclear cells) from LoPFEC or LoKAT (Low Post-Weaning Fecal Egg Count) Katahdin ewes, but these levels return to similar amounts as those observed in HiPFEC or HiKAT (High Post-Weaning Fecal Egg Count) Katahdin ewes by 6 hours of stimulation. This pattern suggests that while TNFα initially responds more strongly in LoKAT ewes, the response normalizes over time, which is ideal as prolonged inflammation can lead to chronic conditions.
IL-1β (Interleukin-1 Beta) is a key pro-inflammatory cytokine that plays a central role in the immune response by promoting inflammation and fever. It is often produced in response to infections and injury, helping to activate other immune cells and drive the inflammatory response. In the results, IL-1β is upregulated early in LoKAT sheep, similar to TNFα, suggesting that PBMCs from these sheep recognize LPS more rapidly. Interestingly, HiKAT sheep show the highest IL-1β expression at six hours post-LPS exposure, indicating a delayed but sustained response and production of inflammatory mediators to LPS. This suggests that while LoKAT sheep exhibit a rapid inflammatory response, HiKAT sheep may have a slower, more prolonged activation of their inflammatory response.
IFN-γ (Interferon Gamma) is a crucial cytokine in the immune system, primarily driving TH1 responses and enhancing cellular immunity. It is vital for activating macrophages and modulating immune responses between TH1 and TH2 pathways. In LoKAT sheep, IFN-γ levels are transiently elevated at 6 hours post-LPS exposure, indicating a strong initial immune response that quickly downregulates, suggesting efficient resolution of inflammation. In contrast, HiKAT sheep show peak IFN-γ upregulation at 9 hours with sustained elevation, reflecting a prolonged or persistent immune response. This indicates ongoing inflammation and a continuous role for IFN-γ in maintaining a proinflammatory state.
While the data is preliminary, and it is too early to definitively claim that Low-PFEC Katahdin sheep are healthier in the face of bacterial insults and require fewer antimicrobial treatments, several important observations can be made. The preliminary findings suggest that pro-inflammatory cytokine expression occurs more rapidly in Low-PFEC Katahdin sheep. Additionally, mediators of inflammation such as TLR4 and NLRP3 are more readily upregulated during LPS stimulation in Low-PFEC Katahdin sheep. This upregulation indicates a greater likelihood of LPS recognition and binding, which may contribute to reduced symptoms of illness. These insights provide a foundation for further investigation into the health benefits associated with Low-PFEC Katahdin sheep and their potential for improved disease resistance.
Education & outreach activities and participation summary
Participation summary:
Outcomes from this project were presented to sheep producers nationwide through various media and events, with a specific focus on Katahdin producers. Research results were shared with producers at several key events. Regionally, the Virginia Tech Southwest Virginia AREC Sheep Field Day and Ram Test Sale provided an ideal venue for disseminating our findings. Annually, over 150 producers attended this educational event, and presentations were recorded and made available online. To further regional outreach, we presented our data at the Eastern Alliance for Production Katahdin annual meeting, which included 50-75 progressive, research-minded producers interested in the latest advancements for improving their flocks.
Furthermore, we plan to submit the final manuscript to Translational Animal Science or The Journal of Animal Science for publication, reaching small ruminant researchers across the country. To directly communicate with producers, we also plan to write a short, accessible publication for a newsletter format. This newsletter, will be published in The Katahdin Hairald Magazine, and serves as a direct communication channel with small ruminant producers about the benefits of selecting for PFEC EBV.
Project Outcomes
The research project focusing on the differential immune response of LoPFEC and HiPFEC sheep to lipopolysaccharide (LPS) stimulation holds promise for positively impacting agricultural sustainability. The early indications of enhanced pro-inflammatory cytokine expression and toll-like receptor 4 (TLR-4) upregulation in LoPFEC Katahdins suggest potential benefits for small ruminant producers. If further studies confirm these findings, it could translate into economic advantages through improved livestock health, reduced antibiotic usage, and lowered veterinary costs. Additionally, the prospect of breeding programs guided by the identified genetic markers could contribute to long-term sustainability by fostering resilient and disease-resistant livestock. Environmentally, reducing chemical inputs, such as antibiotics, aligns with sustainable farming practices, minimizing the environmental impact of agriculture. Additionally, the emphasis on animal welfare and sharing knowledge within the livestock producer community can foster positive consumer perceptions and contribute to a more sustainable and ethical approach to large-scale animal agriculture. While this initial research doesn’t provide conclusive evidence, it is undoubtedly a positive step toward enhancing the health of small ruminants.
The first objective characterizes the differential immune response between LoPFEC and HiPFEC sheep to lipopolysaccharide (LPS) stimulation in-vitro. It has shown promising results with LoPFEC Katahdins upregulating gene expression of pro-inflammatory cytokines (TNF-a, IL1B, and IL-4) within 3 hours of LPS stimulation. The toll-like receptor 4 (TLR-4), responsible for LPS recognition and binding, was also shown to be significantly upregulated within the LoPFEC Katahdins, indicating that these sheep may potentially be able to recognize and bind LPS earlier than HiPFEC individuals. From this data, a 3-hour stimulation time point was selected to explore differences in response to LPS activation from a transcriptome-wide approach. To determine transcriptome-wide gene expression differences between LoPFEC and HiPFEC-derived cells stimulated with LPS, 5 sheep from each genotype were randomly selected so that a total of 20 RNA-seq analyses were performed (10 LoPFEC; 5 Control & 5 LPS-treated and 10 HiPFEC; 5 Control and 5 LPS-treated). This RNA has been submitted to the WVU Genomics Core for cDNA library build and RNA sequencing, and we are awaiting those results to submit a manuscript.