Final report for GNE22-286
Project Information
This project identified associations between DNA methylation and disease status and how these associations may influence performance in dairy cattle. Disease is costly to the dairy industry and negatively impacts animal welfare. Mastitis alone is estimated to cost the U.S. industry $2 billion annually. Genetic selection for disease resistance is possible but the trait is largely impacted by environmental factors that modulate gene expression through epigenetic mechanisms such as DNA methylation. This project consisted of a whole-herd retrospective analysis of two herds, the Cornell Veterinary School Teaching Dairy (VTD) and Cornell University Ruminant Center (CURC), and a DNA methylation analysis on a subset of animals. Performance outcomes of offspring such as production, fertility, health, and longevity were compared based on their dam’s history of disease using electronic records from the farms. Regions of differential methylation between heifers pre- and post-ketosis events and in offspring with differing dam metritis history were identified to characterize a possible biologic explanation for observed associations in health and performance. This project is a pilot project for a more extensive study of epigenetic associations with disease and development in dairy cattle. Understanding the lasting and transgenerational effects of disease can help producers make educated management decisions. Characterizing DNA methylation as a source of phenotypic variation will also help improve genetic prediction models. These two factors will increase farmer profitability with improved selection and management of dairy cattle.
This project consisted of two methodologies: a retrospective whole-herd record analysis and DNA methylation sequencing on a subset of animals. First, an analysis of the Cornell Veterinary School Teaching Dairy (VTD) and Cornell University Ruminant Center (CURC) historical health and performance records was conducted to validate the results of the Carvalho study in this herd i.e., calculate associations between maternal disease status and offspring disease risk and performance. Second, genome-wide DNA methylation analyses of a subset of animals from CURC was conducted to investigate epigenetic mechanisms underlaying the relationships observed in the first analysis. Differences in DNA methylation between sick and healthy animals pre- and post-disease event, whether these are inherited by offspring, and whether these methylation differences have effects on health, growth, production, fertility, and longevity was investigated.
Specific objectives within the study to meet our goal of improved understanding of the relationship between DNA methylation, disease, and phenotypic variation were as follows.
- Objective 1: Investigate associations between dam history of disease and offspring’s susceptibility to disease, production, fertility, longevity, and genetic merit using a retrospective study design of the VTD and CURC
- Objective 2: Characterize associations between DNA methylation and disease status
- Objective 3: Characterize DNA methylation inheritance with a focus on dam disease status and the association with offspring disease susceptibility
- Objective 4: Identify differentially methylated regions with putative biological functions relevant to health, growth, production, fertility, and longevity relationships observed
The purpose of this project was to identify associations between DNA methylation and disease status and how these associations may influence performance. Disease negatively impacts animal welfare and reduces farmer profitability through treatment costs, lost production, and involuntary culling. Genetic evaluations for many diseases are available in dairy cattle allowing producers to select for disease resistance and improve the overall profitability of their animals. However, the direct genetic effect on disease susceptibility is complicated by large environmental and management effects. Epigenetics are a collection of biologic mechanisms used to modulate gene expression in response to environmental changes. A specific example of this is DNA methylation which is a modification of DNA that regulates a gene’s expression. In livestock, biological and environmental factors such as nutrition, stress, and disease have been shown to cause changes in DNA methylation patterns. Furthermore, individual differences in DNA methylation patterns have been associated with phenotypic variation in milk production and immunity. There is even growing evidence that epigenetic marks can be inherited or programmed in-utero based on a dam’s environment affecting the expression of an offspring’s genes throughout their lifetime. We will examine how historic health events in a dam may influence the inheritance of DNA methylation patterns to their offspring and how offspring’s health events may change their own methylation patterns and affect their performance.
Recent studies supporting these concepts include results showing cows at lower risk for clinical disease if their dam’s experienced clinical disease before or during gestation (Carvalho, et al., 2020). Additionally, studies have found that dams under metabolic stress have calves with DNA methylation patterns supporting better energy efficiency than those born to non-stressed mothers (Chaput & Sirard, 2020; Zhang, et al., 2021). This suggests the dam and offspring undergo epigenetic reprogramming to better deal with maternal stressors and disease.
This project consisted of two methodologies: a retrospective whole-herd record analysis and DNA methylation sequencing on a subset of animals. First, an analysis of the VTD and CURC historical health and performance records was conducted to validate the results of the Carvalho study in these herds i.e., calculate associations between maternal disease status and offspring disease risk and performance. Second, genome-wide DNA methylation analyses of a subset of animals form CURC was conducted to investigate epigenetic mechanisms underlying the relationships observed in the first analysis. Differences in DNA methylation between sick and healthy animals pre- and post-disease event, whether these are inherited by offspring, and if these methylation differences have effects on health, growth, production, fertility, and longevity was investigated. This project is intended as a preliminary investigation informing the direction of a larger study of epigenetic inheritance and epigenetic variation in dairy cattle. The goal is to improve the scientific community’s understanding of phenotypic variation and improve genetic prediction models. Improved genetic selection resources and a better understanding of the lasting effects of disease will support farmer profitability in the future.
Research
Proposed Materials and Methods:
Animals and Performance Data: The proposed project will use Holstein at the VTD which houses 175 lactating/dry cows, births 110 female calves, and raises 80 heifers annually. While this is a teaching dairy, it is operated as a commercial dairy herd that markets milk through a cooperative. Cows average 85 lbs/day and are housed in a free-stall facility. Electronic records of birth, breed, production, breeding, health, and culling are regularly maintained in DairyComp305 and will be used in this study to classify disease status and performance outcomes. The VTD has a 12% incidence of metritis, 6% retained placenta, 13% ketosis, 1% milk fever, 2% displaced abomasum, and 27% incidence of mastitis. All animals in the herd are genotyped and have genomic evaluations available that will be used to compare genetic merit. As part of this study, additional performance data that is not routinely collected by the farm will be recorded with help from our research staff moving forward. This includes monthly weight collections and weekly body condition scores as heifers and calfhood disease incidence such as pneumonia and scours. Data will be collected by farm and research staff as appropriate, and procedures will follow IACUC protocols.
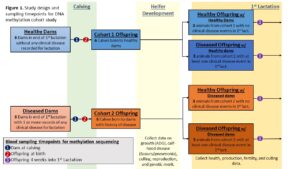
Cohort Definitions and Study Design: This project relies on two methodologies: a retrospective whole-herd record analysis and DNA methylation sequencing on a subset of animals. In both methods, the study subjects will be assigned to two cohorts based on their dam’s incidence of disease in the lactation preceding their birth. Any animal whose dam had at least one recorded incidence of metritis, retained placenta, ketosis, milk fever, displaced abomasum, or mastitis will be assigned to the ‘diseased dam’ cohort and any animal whose dam has no record of any of the previously mentioned or other diseases will be assigned to the ‘healthy dam’ cohort. Due to cohort definitions, only animals born to multiparous dams will be included in the analysis. For the first methodology, differences in first lactation performance and health will be analyzed between cohorts utilizing historic records (Obj. 1). For the second methodology, a subset of the herd will be enrolled in two cohorts (6 offspring and their respective dams each) and differences in DNA methylation will be analyzed between cohorts (Obj. 2-4). The study design for the second methodology is illustrated in the attached figure.
Biological Samples and Epigenetic Data: Biological samples will only be utilized for the second methodology of the study. As part of our larger investigation, all female calves born at the VTD and their dams will undergo routine blood sample collections within 24 hours of parturition starting in May 2022. DNA will be extracted from these samples and frozen. This will create a library of samples for the larger project and allow enrollment into this project based on both dam and offspring disease status. For this project, 12 study subjects and their dams (n=12 offspring; 12 dams) will be used. Enrollment will be dependent on both the dam’s history of disease and the offspring’s disease incidence within the first 4 weeks of their 1st lactation. We will have 3 subjects from ‘diseased dams’ who also had their own disease event, 3 from ‘diseased dams’ without their own disease event, 3 from ‘healthy dams’ with their own disease event, and 3 from ‘healthy dams’ without disease. This sampling strategy is illustrated in the submitted figure. For the enrolled animals, an additional blood sample will be taken 4 weeks into their 1st lactation (n = 12 offspring) following IACUC protocols. The genome wide methylation profile will be analyzed via reduced representation bisulfite sequencing (RRBS). RRBS is cost effective as only fragments pertaining to CpG-rich regions of the DNA (1-5% of genome, mostly promoter regions) are sequenced. Hyper and hypo methylated regions and differentially methylated regions (DMR) between groups will be identified. Gene ontology and KEGG pathway enrichment analysis will be performed to functionally characterize the differentially methylated regions and genes in terms of what is known about the biological function of genes.
Objective 1: Investigate associations between dam history of disease and offspring’s susceptibility to disease, production, fertility, longevity, and genetic merit using a retrospective study design of the VTD.
Performance Outcomes: For this objective, the whole herd will be split into ‘diseased dam’ and ‘healthy dam’ cohorts to analyze differences in 1st lactation health and performance of their respective offspring. The presence or absence of any clinical disease and the number of disease events in 1st lactation will be used as health outcomes in the analysis. Total 1st lactation milk, fat, and protein production, heifer and 1st lactation insemination and pregnancy rates, and culling rate and reasons will be used as performance outcomes in the analysis. Additionally, the difference between genomic predicted production and realized first lactation production will be used as an outcome to be compared between cohorts in order to assess if maternal disease status may contribute to phenotypic variability from genomic predictions.
Statistical Analysis: Analyses similar to the Carvalho study will be used to analyze differences in offspring health and performance between cohorts using R version 4.1.1 (Carvalho et al., 2020). ANOVA will be used for continuous outcomes, logistic regression will be used for binary outcomes, and Cox proportional hazard regression will be used for intervals to events. Data such as season, age, and parity of the data will be collected and assessed as covariates in the model.
Objective 2: Characterize associations between DNA methylation and disease status.
Dam Disease Comparison: Differences in DNA methylation will be identified between dams with and without a history of clinical disease based on the samples collected at calving. Differentially methylated regions (DMR) between the diseased and non-diseased dams could stem from two possible options. One, these DMRs could indicate differences existing before the disease incident that contributed to either their susceptibility or resistance to disease or the DMRs could be differences the animals acquired in response to the disease.
Pre- and Post- Offspring Disease Comparison: In order to determine the source of the DMR found between dams, methylation patterns for individual offspring at birth will be compared to their methylation patterns 4 weeks into their first lactation and to their dams. The peak incidences of all the diseases of interest are within the first 4 weeks of lactation. Therefore, changes in methylation between birth and lactation present only in offspring that had their own disease event, not in offspring that did not have disease, could be considered changes in methylation as a response to disease. In contrast, DNA methylation patterns of offspring at birth in relation to whether they end up having a disease event in their first year of life, suggests DNA methylation provides either resistance or susceptibility to the disease. These DMRs identified in the offspring at birth or as a response to disease will be compared to the patterns found in the dams.
Objective 3: Characterize DNA methylation inheritance with a focus on dam disease status and the association with offspring disease susceptibility.
Offspring Cohort Comparison: DMR in offspring at birth will be identified between cohorts of dam disease status. DMR identified here would represent differences in methylation in offspring due to maternal disease status near gestation. The diseases studied in this project are most common in early lactation meaning they occur before or around the time of insemination. The DMR between the two cohorts of offspring would have either been the result of in-utero programming of the offspring’s methylation pattern due to its own exposure to the disease or inherited from the dam based on her methylation pattern. The analysis for objective 1 investigated the source of the methylation patterns of the dam.
Inheritance Comparison: The DMR found between the two cohorts of offspring will be compared to the DMR between sick and healthy dams. If similar regions are found, this supports that either offspring and dam experienced the same re-programming event as a response to their exposure to the disease or the dam passed her methylation pattern on to the offspring. Without more frequent sampling, it will be hard to determine which scenario is the case, but we will know the outcome relationship between offspring methylation and disease.
Objective 4: Identify differentially methylated regions with putative biological functions relevant to health, growth, production, fertility, and longevity relationships observed
Performance Analysis: A similar performance outcome analysis (excluding disease incidence) as conducted for objective 1, will be conducted for the 12 methylation study subjects. While the power of this analysis will be much lower than the whole-herd analysis, it will be conducted to assess whether the direction of trends is the same in the subset of animals as in the whole-herd. Heifer average daily gain and calfhood disease incidence will be included as additional performance outcomes. This is not something that is routinely collected by the farm but will be collected for the study subjects of this project. Previous studies that investigated the impact of maternal disease status on offspring performance, proposed poor heifer growth as a possible reason for the increased cull rate of heifers born to diseased dams but did not collect the data to investigate that claim (Carvalho et al., 2020).
Functional Analysis: Genes near the identified DMRs in Objectives 2-4 will be identified. RRBS focuses on methylation patterns near promoters which are regions that impact gene expression. Genes found near DMR could indicate differentially expressed genes between groups. Gene ontology and KEGG pathway enrichment analysis will be conducted to functionally categorize the nearby genes. Gene ontology and KEGG pathway analyses use databases having genes cross-referenced with functional research to provide annotation as to what the biological function is of a gene. This should reveal genes that have biological functions that support the phenotypic relationships between maternal disease status and offspring health and performance previously identified (in the whole herd and subset).
Progress Report Methods Adjustments:
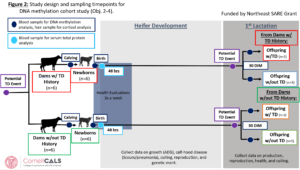
Methods adjustments (January 2023): Since beginning the study, a few adjustments have been made to the protocol. Health evaluations for calves have been implemented as a way of monitoring calf health between birth and weaning. These are conducted three times a week and include rectal temperatures and eye, ear, nose, cough, fecal, and body condition scores. An additional blood sample timepoint was added at 48 hours after birth. This sample is used for a serum total protein estimation using a digital refractometer. Serum total protein is a good estimator of serum IgG levels which are indicative of the success of passive transfer of immunity through colostrum. As failure of passive transfer could be a confounder for future growth, disease susceptibility and productive outcome, we can now control for this in future models. This change was implemented on 7/1/22. The first 4 calves enrolled in the study do not have serum total protein readings. Due to logistical concerns with the scale, bodyweights are now being estimated using a weigh tape once a week during the time from birth to weaning. This change was implemented on 9/21/22. The 16 calves born since implementation and all future calves will have complete pre-weaning growth information. Enrollment of bull calves has stopped as of 9/14/22. An updated version of the study design schematic has been added as figure 2.
Methods adjustments (January 2024 Report):
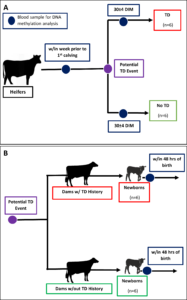
As explained in the previous annual report, we were struggling to enroll multiparous dams with a history of disease at the VTD. To rectify this, we decided to conduct targeted sampling at a different farm, the Cornell University Ruminant Center (CURC). CURC has a larger herd (~500 head) making it easier to find the animals we need. However, this herd requires payment per animal used so we developed a more targeted approach. Figure 3 displays the adjusted study design. In part A, blood samples will be collected from primiparous heifers within the week before calving and then again at 30 DIM with the goal of getting 6 heifers who have a TD event in the first 30 DIM and 6 calving-date matched heifers that do not experience a disease event. All disease diagnosis was done by farm staff and electronically recorded. This will allow us to find DMR in heifers before calving that may be risk indicators for TD by comparing the pre-calving samples from those that got TD to those that did not get TD. We can also identify DMR that may be acquired in the individuals that experience disease by comparing the pre- and post-calving samples of an individual. This supports objective 2 and 4. In part B of Figure 3, 6 cows due with their second calf that had a TD event in their first lactation and 6 due-date and parity matched cows without a history of TD will be identified pre-calving. Blood will be collected from their calves within 48 hours of birth. From these samples we will be able to identify DMR between calves born to dams with a TD history and those born to dams without a history of TD. This applies to objective 3 and 4. Our ability to track these calves’ future progress is limited because we had to enroll male and female calves in order to reach our sample size. All males and some females are sold soon after birth and we will not have the ability to assess their future performance. Of the female calves that were chosen to remain in the herd, we did get an additional blood sample at 48 hours of birth to use for serum total protein analysis, a measure of passive transfer of immunity. We do plan to look at their future performance, but our sample size will be smaller than what we require to draw concrete conclusions.
In the original proposal, we were planning on using reduced representation bisulfite sequencing. We have since decided to do whole genome bisulfite sequencing as the price has reduced since the project proposal was submitted. This will give us whole genome coverage instead of just the CpG-rich regions. This is the gold standard for DNA methylation sequencing.
Final Material and Methods (December 2024):
Animal Records and Analysis:
All current and archived records were extracted from DairyComp for both the VTD and CURC. Records spanned 2012-2024, totaling about 1600 animals from the VTD and 5000 animals from CURC. Performance metrics such as culling, disease, and standardized milk production were pulled for every animal. From birth to first lactation, disease events were grouped into digestive, respiratory, and other disease groups. In 1st lactation, disease events were grouped into uterine, metabolic, digestive, mastitis, and other disease groups. Animals were classified based on both their dam’s parity (Primiparous vs. Multiparous) and their dam’s disease status either in the lactation prior to their birth for Multiparous dams or from the dam’s birth until her first calving for primiparous dams. Offspring were placed in the Dam_D group if the dam had any disease reported regardless of the type of disease and Dam_No_D group, if the dam was not reported to have any type of disease. Linear or logistic mixed effects models were fit for each outcome with fixed effects of dam parity, dam disease status, and their interaction. Farm was always included as a random effect and all combinations of random effects of birthdate year, birthdate group (grouping 3 consecutive birthdate years), sire, and dam were tested. The best model was chosen to minimize AIC. Survival analysis was conducted to analyze the difference in survival probability over the whole lifetime of animals with fixed effects of dam parity, dam disease status, and their interaction and random effects of birthdate group.
Biological Sampling:
Sampling for this project completed in November of 2023. After enrolling 49 Holstein heifer calves at the VTD from July 2022-June 2023 and failing to capture any born to dams with a history of TD an adjusted and more targeted sampling plan was enacted in July of 2023 at CURC. From July-November 2023, blood was collected from 24 primiparous heifers within the week before their first calving and again at 30 DIM. Blood was also collected from 12 calves within 24-48 hours after their birth.
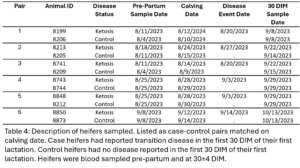
Primiparous heifers underwent farm monitoring and diagnosis of TD during the first 21 DIM. Of the 24 primiparous heifers sampled, 9 experienced at least one instance of TD, specifically there were 7 cases of subclinical ketosis, 2 cases of metritis, and 1 case of clinical mastitis. Farm diagnosis of TD was based on a combination of activity monitor health alerts and clinical sign observation. For ketosis specifically, all fresh cows had blood beta-hydroxybutyrate (BHBA) measured at 3-9 DIM and were classified as healthy (BHBA < 1.3 mmol/L), subclinical ketosis (1.3-2.9 mmol/L), or clinical ketosis (>3.0 mmol/L) cases by farm personnel.
All 12 calves sampled were their dam’s second calf and were selected based on their dam’s history of disease in their first lactation. Five of the case dams had metritis, 1 had subclinical ketosis, and the other 6 control dams had no disease reported in their first lactation. Case and control dams were matched on due date and calves were classified based on their dam’s classification. There were 4 male and 2 female calves within both case and control groups.
Whole blood genomic DNA was extracted from all blood samples collected throughout the project and banked at -80°C.
Sequencing and Epigenetic Data Analysis:
Six case-control pairs of heifers were selected for sequencing from the CURC dataset. All cases selected had ketosis and were matched to the heifer with the nearest calving date and no reported disease. The 6 case-control pairs of calves from CURC were also sequenced. Whole genome bisulfite sequencing (WGBS) for this study was contracted out through Novogene who was responsible for library preparation and sequencing at an estimated 15X coverage on an Illumina Novaseq 6000 sequencer. The msPIPE pipeline was used to conduct quality control via TrimGalore!, create a bisulfite converted reference genome (ARS_UCD1.2/bosTau9), align reads, and call DNAm via Bismark (Kim, et al., 2022; Martin, 2011; Krueger & Andrews, 2011). In R, DMRichR was used to smooth methylation, identify differentially methylated regions, annotate them to functional regions and nearby genes, and run machine learning classification algorithms to identify the most informative DMR (R Core Team, 2021; Laufer, et al., 2020; Korthauer, et al., 2012; Hansen, et al., 2012). Gene Ontology analysis was conducted using GOFuncR and the significance of overlap between differentially methylated gene lists was tested using GeneOverlap in R. Visualizations were made in R.
Progress Report Results:
Biological Samples and Epigenetic Data Progress (January 2023): As of January 6, 2023, 39 Holstein heifer and 5 bull calves have been successfully enrolled in the study marked by successful blood collection from both calf and dam within 24 hours of birth. Of the calves enrolled, 6 heifer calves and all bulls were sold and are no longer part of the study. One heifer calf was euthanized. Of the 32 calves that have remained in the herd, 21 were their dam’s first calf, 10 were born to multiparous dam’s with no history of transition disease in the previous lactation, and 1 was born to a dam previously treated for hypocalcemia. They have been sired by 8 different sires. 7 calves have been treated for scours and 1 was treated for a navel infection. 14 have been successfully weaned and are being transferred to the heifer raiser. The remaining heifer calves are still at the farm and are being monitored for growth and disease events until weaning.
We do have concerns about the lack of calves being born to dams with a history of transition disease. Historically, this has not been a problem, but recently breeding strategies of the VTD herd have placed more emphasis on sourcing replacement animals from primiparous animals. Most heifers are bred with sexed semen; whereas, most multiparous animals are bred with conventional dairy semen or beef semen. Furthermore, the likelihood that an animal is bred with beef semen likely increases if she is a “problem cow” that has a history of disease events. The targeted use of sexed semen and beef semen is a progressive breeding strategy allowing herds to maximize genetic progress and gain a higher sale price for calves not intended as replacements. For this study, it is a problem because there are a small proportion of Holstein heifer calves that could qualify for objectives 2-4. We are considering partnering with a larger commercial herd for sourcing the biological samples for this part of the project.
Tentatively, enrollment of all Holstein heifer calves born at the VTD will continue until June 2023. All calves enrolled will be followed to their first calving (beginning spring of 2024). At 4 weeks into their lactations, we will choose 12 heifers based on their own and their dam’s disease status (as previously described in the proposal). An additional blood sample at 4 weeks into the lactation will be collected from these animals. Birth and lactation blood samples of the 12 selected dams and calves will be analyzed by RRBS DNA methylation sequencing. The sequences will be used for Objectives 2-4.
Objective 1 Progress (January 2023): The dam disease status for all current cows in the herd is being extracted from the herd’s archived DairyComp records. To date, all current first and second lactation cows have been classified as either born to a primiparous dam (excluded from study), born to a dam with no record of clinical disease in the lactation coinciding with their gestation (“healthy dam” cohort), or born to a dam with recorded clinical disease in the lactation coinciding with their gestation (“diseased dam” cohort). Dam disease events have included mastitis, metritis, retained placenta, ketosis, displaced abomasum, and hypocalcemia and have not yet been filtered by timing in lactation. First lactation production and disease events have also been extracted for all current first and second lactation animals in the herd. Third and greater lactation animals currently in the herd will be classified by dam disease status and their archived first lactation records will be extracted in the coming months. Statistical analysis of this data will then be conducted to investigate the relationship between dam disease history and offspring performance.
Objective 2 – 4 Progress (Jan 2023 Report): DMR analysis will commence once study subjects have been selected and RRBS sequences have been received (estimated summer 2024).
Biological Samples and Epigenetic Data Progress (January 2024 Report):
All sampling for the adjusted study design has been completed as of November 2023. All heifers due to calve starting in July 2023 were sampled 1x a week starting two weeks before their due date until they calved. They were then sampled again at 30 ± 4 DIM. This was done routinely until the goal of 6 matched pairs were enrolled. In total, 24 heifers were enrolled making eight case-control pairs. Six of the cases had ketosis, one had ketosis and metritis, and one had metritis. The six pairs with ketosis have been chosen for the study.
The multiparous dam case-control pairs were selected in July 2023 using the farm’s electronic records. Eight were chosen as a precaution in case there were stillborn calves and one calf was stillborn. Of the six pairs of calves successfully enrolled, 1 calf was born to a dam with a history of ketosis and the other 5 case calves had dams with a history of metritis. 8 calves were male and 4 were female. The female calves who were selected to remain in the herd had an additional blood collection between 36-48 hours after birth for serum total protein reading.
DNA has been isolated and stored for all blood samples collected for the project. The samples needed for sequencing have been selected (36 in total) and were sent to Novogene for sequencing in early January.
Final Results and Discussion (December 2024):
Objective 1: Investigate associations between dam history of disease and offspring’s performance using a retrospective study design
Descriptive Statistics:
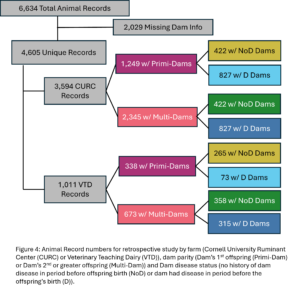
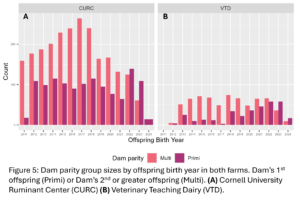
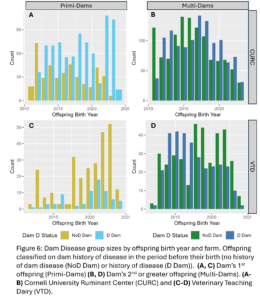
A total of 4,605 unique offspring records were extracted from electronic records of both farms after removing animals who did not have a dam listed in the records or who had a dam who could not be classified by her disease status (Fig. 4). These records spanned 12 years and overall CURC had more animal records than the VTD. In both farms there were more offspring from multiparous dams than primiparous dams in total, but the number of offspring from primiparous dams increased in recent years in both farms (Fig. 5). In the CURC dataset, there were more offspring from dams with a history of disease in total and that seems to be due to an increase in offspring from these dams in more recent years (Fig. 6). In the VTD dataset, the opposite was true; overall there were less offspring to dams with a history of disease and there was a decreasing trend in the number of offspring from diseased dams in recent years (Fig. 6).
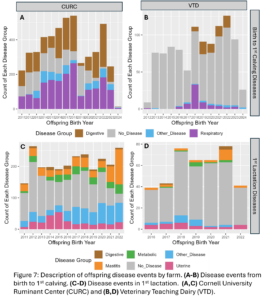
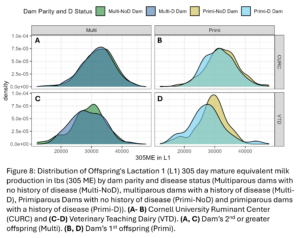
Disease outcomes were separated into two timeframes for assessing offspring performance: from birth until 1st calving and in her 1st lactation. Disease events were grouped into digestive, respiratory, and other diseases from birth to 1st calving and into digestive, metabolic, uterine, mastitis, and other diseases within 1st lactation. The number of cases of each disease type fluctuated from year to year and across farms (Fig. 7) but overall, there was a total disease prevalence of 57% from birth to 1st lactation and 44% in 1st lactation. Culling and milk production were also evaluated as offspring performance outcomes. Overall, 35% of animals left from birth to 1st lactation and 13% of animals that made it to 1st lactation left in 1st lactation. Standardized 305 day mature equivalent milk production was numerically higher in CURC animals than VTD animals (Fig. 8).
Effect of Dam Parity on Offspring performance:
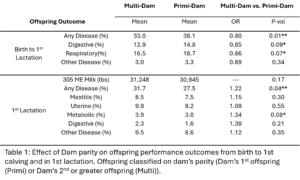
All animals were classified based on their dam’s parity. All animals who were her dam’s first calf were in the Primi-Dam group and animals who were her dam’s second or greater calf were grouped in Multi-Dam. The effect of dam parity on offspring milk production and disease risk was evaluated (Table 1). The calves from the Multi-Dam group were less likely to have disease (OR = 0.80, p-value = 0.02), specifically respiratory disease (OR = 0.86, p-value = 0.07) from birth to first lactation than the calves from the Primi-Dam group. Of the offspring that made it to first lactation, offspring of multiparous dams were more likely to have disease (OR = 1.22, p-value = 0.04) specifically metabolic disease (OR=1.34, p-value=0.08). There was no significant difference in milk production between offspring of differing dam parities. There was no significant difference in lifetime survival probability between Multi-Dam and Primi-Dam offspring.
Effect of Dam Disease History within dam parity groups:
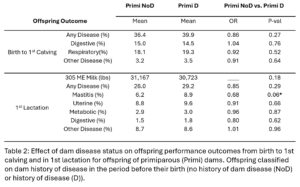
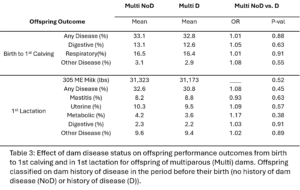
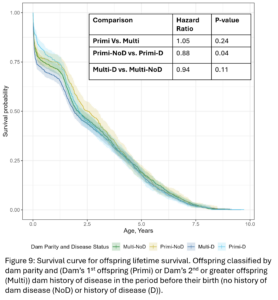
Within dam parity groups, all animals were classified based on their dam’s disease history in the period preceding their birth. Within the Primi-Dam group, the offspring was classified based on the dam’s disease records from her birth to her 1st calving and placed in the Primi D if there was any disease reported or in Primi NoD if there was no disease. Within the Multi-Dam group, offspring were placed in the Multi D group if the dam had any disease reported in the lactation before the offspring was born or Multi NoD if there was no disease reported in that lactation. The effect of dam disease status on offspring milk production and disease risk was evaluated (Table 2 and 3). There were no significant differences between Primi D and NoD offspring from birth to first calving. In first lactation, Primi NoD offspring tended to be less likely to have mastitis (OR=0.68, p-value = 0.06). Across their whole lives, offspring of Primi NoD were at less risk of leaving the herd than offspring of Primi D (HR=0.88, p-value = 0.04; Fig. 9). There were no significant differences in disease, milk production, or culling outcomes between Multi D and Multi NoD offspring.
Discussion and Limitations:
In our study we saw a bigger effect of dam parity on offspring performance than dam disease status. Similarly to Carvalho, et al. we saw offspring of multiparous dams were more likely to have disease in 1st lactation than offspring of primiparous dams and that there was no significant effect of dam disease status on offspring milk production. Carvalho, et al. observed a lower risk for disease in offspring whose dam had a history of disease. Based on the Carvalho Study, we hypothesized that there was a protective effect of dam disease history on offspring disease susceptibility. Overall, our study does not support this hypothesis. Other than the trend for Primi NoD offspring to have less mastitis than Primi D offspring, we did not see an effect of dam disease history on offspring disease risk. Primi D offspring also had a lower risk of leaving the herd over their whole lifetime than Primi NoD offspring. The Carvalho study did not evaluate the effect of dam disease in primiparous dams. Variation in disease diagnosis and monitoring across years, personnel, and farms was hard to completely control for in the analysis. This is likely contributing to the variation between our results and previously published studies and makes studies like this difficult.
Objective 2: Characterize associations between DNA methylation and disease status
Description of Animal Characteristics and Global Methylation:
The pre-calving and 30 DIM samples from six case heifers with subclinical ketosis (TD) and their matched healthy controls (NoTD) were chosen for WGBS. Cases all with the same transition disease were chosen to minimize variation as it would be expected that different transition diseases would have different epigenetic effects. The disease status, calving date, disease date, and sampling dates are displayed in table 4 and case control pairs are listed together.
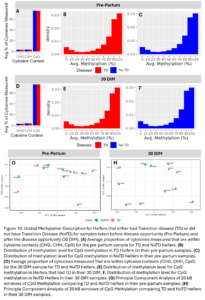
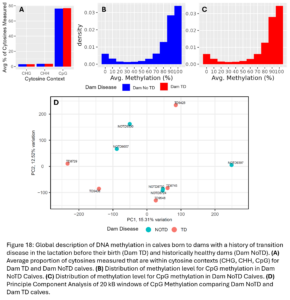
Across the 24 samples (12 animals at 2 time points), we identified 3,511,854 cytosine sites. The majority of them were CpG sites (76%) and 3.1% and 3.5% were CHG and CHH sites, respectively (Fig. 10A and D). Of the CpG methylation sites, most were either highly or lowly methylated and there were not any large differences in the distribution of genome methylation levels between TD and NoTD or between pre-partum and 30 DIM samples which is as expected (Fig. 10B-C, E-F). After smoothing across CpG methylation levels in 20kb windows, there were no large global differences in CpG methylation between TD and NoTD Heifers at either sampling time point (Fig. 10G-H). This is to be expected as all samples are from the same tissue type, blood, and it is more likely that TD would affect DNAm of a smaller number of specific sites.
Possible TD Risk or Protective DNAm loci:
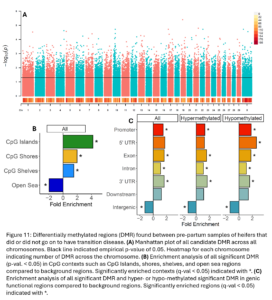
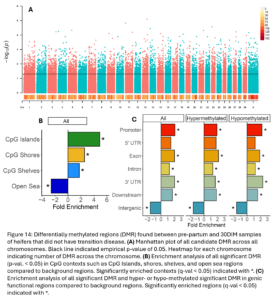
Across the genome, 15,004 DMRs were identified (empirical p-value <0.05) between heifers that went on to have transition disease (TD) and healthy heifers (NoTD) in their pre-partum blood sample (Fig. 11A). Of the significant DMR, 64% were hypermethylated and 36% were hypomethylated in TD heifers versus NoTD heifers. DMR were significantly enriched in CpG islands, shores, shelves, and open sea contexts (Fig. 11B). They were also enriched in promoters, exons, introns, 3’ UTR, and intergenic functional regions. Hypomethylated DMRs were also enriched in 5’UTR (Fig. 11C).
Two machine learning algorithms were used to classify TD and NoTD heifers based on DMR identified pre-partum. There were 33 DMRs that were ranked important by both algorithms (Fig. 12). Only five of these DMR were hypomethylated and 28 were hypermethylated in TD versus NoTD heifers. DMR were annotated to mostly intergenic regions but 2 were annotated to promoters, 3 to exons, and 4 to introns. These DMR could represent potential risk/susceptibility loci for ketosis as all TD heifers had ketosis specifically. These regions will be characterized further in Obj. 4.
DNAm changes during the transition period:
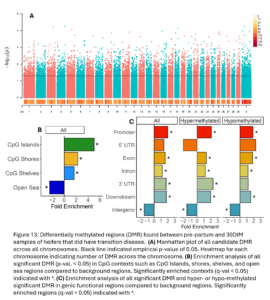
Across the genome, 7,624 DMRs were identified (empirical p-value <0.05) between the pre-partum and 30 DIM samples of TD heifers (Fig. 13A). These DMR were significantly enriched in CpG islands, shores, shelves, and open sea contexts (Fig. 13B). They were also enriched in promoters, exons, introns, downstream, and intergenic functional regions. Hypermethylated DMRs were also enriched in 3’UTR (Fig. 13C).
Across the genome, 8,667 DMRs were identified (empirical p-value <0.05) between the pre-partum and 30 DIM samples of NoTD heifers (Fig. 14A). These DMR were significantly enriched in CpG islands, shores, shelves, and open sea contexts (Fig 14B). They were also enriched in promoters, exons, introns, 3’UTR, downstream, and intergenic functional regions (Fig 14C).
Two machine learning algorithms were used to classify pre-partum and 30DIM samples of TD heifers. There were 19 DMRs that were ranked important by both algorithms. Nine were hypomethylated and 10 were hypermethylated in 30 DIM samples compared to Pre-Partum samples (Fig. 15). These DMR were annotated to mostly intergenic regions but 2 were annotated to promoters, and 2 to introns.
Two machine learning algorithms were used to classify pre-partum and 30DIM samples of NoTD heifers. There were 24 DMRs that were ranked important by both algorithms (Fig. 16). Half the DMR were hypomethylated and half were hypermethylated in 30 DIM samples versus pre-partum samples. These DMR were mostly annotated to intergenic regions but two were annotated to promoters, one to an exon, and six to introns.
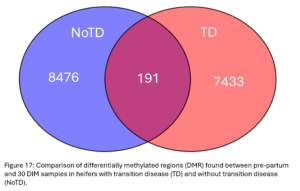
DMRs identified between pre-partum and 30 DIM samples represent the changes in DNA methylation patterns animals experience in the transition period. DMR identified between these points in NoTD animals likely represent the changes healthy animals go through over this time and DMR within TD animals could represent the changes sick animals go through. There were 191 regions where both groups experienced DNAm changes (Fig. 17). The 7,433 DMR only seen in TD heifers could represent changes specifically related to their case of ketosis. The 8,476 DMR only seen in NoTD heifers could represent normal changes in DNAm that are prevented in animals with transition disease. These regions will be characterized further in Obj.4.
Objective 3: Characterize DNA methylation inheritance with a focus on dam disease status and the association with offspring disease susceptibility
Description of Animal Characteristics and Global Methylation:
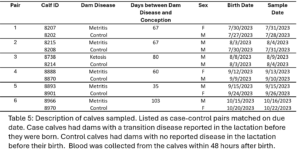
All 12 calves sampled were sent for WGBS. The dam disease status, sex, birthdate, and sampling date are displayed in table 5.
Across the 12 samples, we measured on average 3,263,675 cytosine sites and 77% of them were in CpG contexts, 3.1% in CHG contexts and 3.6% in CHH contexts (Fig. 18A). Of CpG sites, most were either were highly or lowly methylated (Fig. 18B-C) which is as expected. After smoothing across CpG methylation levels in 20kb windows across the genome, there were no large global differences in CpG methylation between Dam_TD and Dam_NoTD calves as shown by the PCA plot (Fig. 18D). We did not expect dam TD to have a large global effect on DNAm of her future offspring. It is more likely there are smaller changes.
Offspring DNAm differences associated with Dam Disease Status:
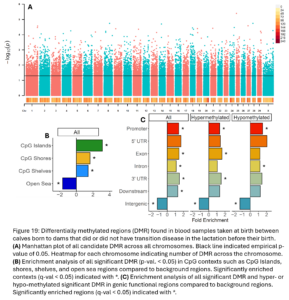
To identify smaller differences in DNAm patterns between case and control calves, differential methylation analysis was conducted to compare 5 case calves whose dam experienced metritis in the lactation before their birth to their 5 matched control calves who had historically healthy dams. For this analysis, the calf whose dam had ketosis and his matched control were removed. This allowed us to have a more targeted analysis on understanding the effects of metritis. We expect different transition diseases could have different epigenetic programming effects. Overall, we identified 12,941 DMRs between calves whose dams did or did not have metritis (empirical p-value <0.05; Fig. 19A). The DMRs were distributed across the genome and 41% were hypomethylated and 59% were hypermethylated in Dam TD calves. DMR were enriched in CpG Islands, shores, shelves, and open sea contexts (Fig. 19B). Hypermethylated DMR were enriched in promoters, exons, 3’UTR, and intergenic regions. Hypomethylated DMRs were enriched in promoters, introns, and intergenic regions (Fig. 19C).
Machine learning algorithms were used to identify the most informative DMR to classify Dam TD and Dam NoTD calves. Nineteen DMR were commonly picked as informative by both machine learning algorithms. Of those, 4 were hypomethylated and 15 were hypermethylated in TD calves (Fig. 20). Most of these DMR were annotated to intergenic regions but one falls within a promoter and 2 within introns.
Objective 4: Identify differentially methylated regions with putative biological functions relevant to health, growth, production, fertility, and longevity relationships observed
Enrichment and Functional Analysis of TD Risk Loci:
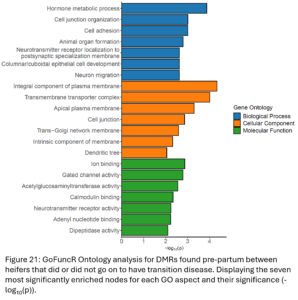
DMRs were annotated to the nearest gene resulting in 5,763 differentially methylated genes (DMG) between TD and NoTD heifers pre-partum. Gene ontology (GO) analysis was run with all of these DMGs (Fig. 21). Enriched biological process GO terms included hormone metabolism and cell junction and adhesion. Enriched cellular component and molecular function GO terms also point to possible differences in cell signaling and transport between TD and NoTD heifers pre-partum.
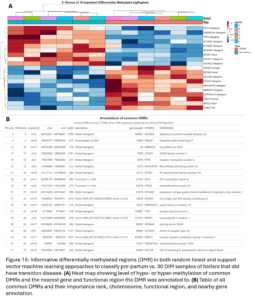
Of all the DMG identified between TD and NoTD heifers pre-partum, 594 were genes in which the DMR fell within a promoter. On chromosome 10, one DMR that was identified as an important DMR through both machine learning algorithms falls within the promoter of the TSHR gene (Fig. 12). This gene codes for thyroid stimulating hormone receptor and thyroid activity differences in cattle with ketosis have been previously reported (Pehrson, et al., 1966). This promoter is hypermethylated in TD heifers suggesting a reduction in the expression of TSHR. Another important DMR from the machine learning classification falls within the promoter of the VCAN gene on chromosome 7 (Fig. 12). This gene codes for versican which is an extracellular matrix proteoglycan important in cell adhesion. In cattle, VCAN has been previously associated with oocyte number, embryo quality, and body measurements (Rocha, et al., 2024; Yan, et al. 2024).
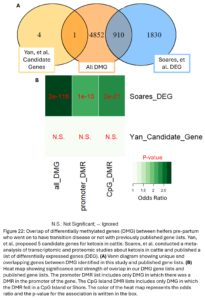
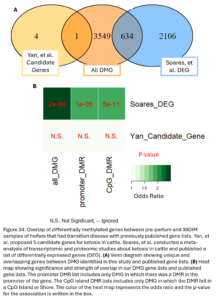
Lastly, we wanted to compare our list of DMG’s to other genes previously implicated in ketosis. Soares, et al. previously conducted a meta-analysis of transcriptomic and proteomic studies about ketosis and negative energy balance in cattle (2021). They published a list of 2,740 differentially expressed genes (DEG) from transcriptomic studies. Yan, et al. conducted a multi-omics study related to ketosis in cattle and proposed a list of 5 candidate genes (2020). Our DMG were significantly overlapped with the Soares study genes. This means we identified changes in DNAm in and around genes that have been previously shown to be differentially expressed during ketosis (Fig. 22B). One DMG, GRINA, was also a candidate gene from the Yan study. This gene has also been previously associated with milk protein percent in cattle (Pedrosa, et al., 2021). This supports that the differential methylation seen between our TD and NoTD heifers pre-partum could be associated with their risk for ketosis and provides more support for the association of these genes with the disease.
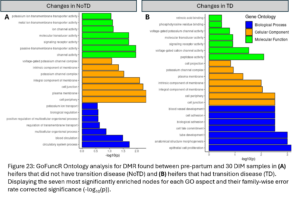
Enrichment and Functional Analysis of acquired DNAm changes with TD:
Of the DMR representing DNAm changes in the transition period only seen in NoTD heifers, 4,525 DMG were identified. Gene ontology analysis was run with all these DMGs (Fig. 23A). Enriched biological processes were related to the circulatory system. The DMG identified may be associated with changes in blood cell proportions during the transition period which has been previously demonstrated (Sun, et al., 2022). Of the DMR representing DNAm changes in the transition period only seen in TD heifers, 4,184 DMG were annotated. Gene ontology analysis was run with all these DMGs (Fig. 23B). Enriched biological processes included epithelial cell proliferation and anatomical structure morphogenesis. Enriched molecular functions included peptidase activity and voltage-gated cation channel activity.
Of all the DMG identified between pre-partum and 30 DIM samples in TD heifers, 429 were genes in which the DMR fell within a promoter and 186 were genes in which the DMR fell within a gene body (exon or intron). NPSR1 was a DMG found between TD heifers pre-partum and at 30DIM and was not seen as differentially methylated in NoTD heifers over those timepoints. A DMR within an intron of the gene was also ranked with high importance for both machine learning algorithms (Fig. 15). This gene codes for a neuropeptide receptor and has been associated with fertility traits in cattle (Jiang, 2019).
Of all the DMG identified between pre-partum and 30 DIM samples in NoTD heifers, 502 were genes in which the DMR fell within a promoter and 256 were genes in which the DMR fell within a gene body (exon or intron). A DMR within the promoter of NMRAL1 was ranked with high importance for both machine learning algorithms (Fig. 16). NMRAL1 codes for a NADPH sensor protein. In NoTD heifers, the promoter of NMRAL1 was hypermethylated at 30DIM compared to Pre-Partum suggesting a decrease in expression in healthy cattle over this time. NAD+/NADPH balance has been shown to be affected by ketogenic diets in humans and NADPH oxidase-ROS signaling pathway is activated in cows with subclinical ketosis (Elamin, et al., 2021; Song, et al., 2022). It is possible the normal increase in DNAm of the NMRAL1 promoter is prevented by a ketosis event explaining why this was not identified as a DMR in TD heifers across the same timepoints.
Lastly, we wanted to compare the DMG that undergo DNAm changes in heifers that had ketosis to previously published gene lists associated with ketosis (Fig. 24A). Our DMG significantly overlapped with the Soares study DEG’s and 1 gene, GRINA, was a ketosis candidate gene from the Yan study. The DMR near GRINA found between pre-partum and 30DIM samples of TD heifers was 32 kbp downstream of the transcription start site of the gene in an intergenic region. A DMR between TD and NoTD heifers pre-partum was also annotated to this gene but it was about 20 kbp upstream of the transcription start site. This provides even more evidence supporting the association of GRINA with ketosis in dairy cattle.
Enrichment and Functional Analysis of inherited DNAm differences due to Dam TD:
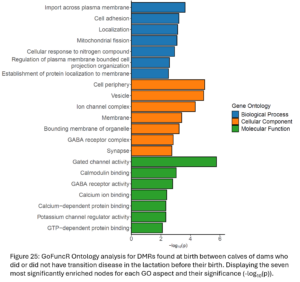
DMRs were annotated to the nearest gene resulting in 5,130 DMGs between Dam TD and Dam NoTD calves. Gene ontology analysis was run with all of these DMGs (Fig. 25). There are biological processes, cellular components, and molecular function GO terms related to cellular membrane transport and signaling.
Of all the DMGs, 1,036 were genes in which the DMR fell within the gene’s promoter. On chromosome 13, one DMR that was ranked as important by both machine learning algorithms falls within the promoter of the CEBPB gene (Fig. 20). The C/EBPβ transcription factor this gene codes for has been previously implicated in epigenetic memory of immune response in hematopoietic stem cells (De Laval, et al., 2020). This gene’s promoter was hypermethylated in TD calves suggesting decreased expression of the gene that could affect the future immune response of calves born to dams with a history of metritis.
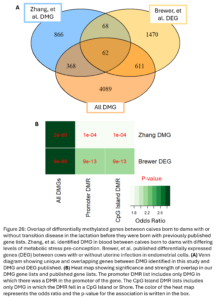
Lastly, we wanted to compare our list of DMG’s to other genes previously implicated in inheritance of maternal transition disease and metritis more broadly. Zhang, et al. previously identified 1,364 DMG’s in calves at birth associated with maternal metabolic stress pre-conception. Metabolic stress is a common cause and outcome of transition disease (2022). Brewer, et al. previously published a list of 2,211 DEG in endometrial cells of cows that had uterine infection compared to healthy cows (2020). Our DMG were significantly overlapped with both gene lists (Fig. 26B). The overlap with the Brewer gene list suggests that calves born to dams with a history of metritis may be inheriting some of the DNAm changes associated with gene expression changes seen in cows who have metritis themselves. The overlap with the Zhang study genes suggests there may be some DNAm changes associated with dam transition disease more broadly that are not disease specific.
Discussion and Limitations:
Overall, we were able to propose three types of DNAm loci associated with transition disease in dairy cattle: potential risk loci, loci that undergo changes in DNAm around the time of disease, and loci in which dam disease status may impact offspring DNAm. There was overlap in the genes that potential risk DMR and DMR undergoing changes due to transition disease were annotated to, though the actual DMR regions were unique. GRINA is an example of one of these genes and is a promising candidate gene for ketosis in cattle. Differential methylation in or around this gene demonstrated in our study may help explain some of the functional connection between this gene and the ketosis phenotype.
There was no overlap in the DMG identified in calves and the DMG identified in heifers in our study. This is likely due to fact that the actual transition disease captured in these populations were different. The dams of our Dam TD Calves had metritis while the TD heifers had ketosis. Nonetheless, the DMR identified between Dam TD and NoTD calves within the promoter of the CEBPB gene is very promising as a potential locus for epigenetic programming of dam disease history. The transcription factor it codes for is known to play a role in immune response and it has been implicated in the epigenetic memory of immune response in hematopoietic stem cells.
While these results are very promising, three limitations of our study should be recognized. Firstly, we only looked at DNAm within one tissue, blood, and did not account for the heterogeneity of cell types within the samples. While the majority of DNA isolated from a blood sample will be from white-blood cells, there are cell-specific DNAm patterns for different white blood cells. We did not account for this and cannot definitively say if the differential methylation we observed is simply from differing blood cell proportions in samples. Secondly, we did not intensively diagnose disease and instead relied on farm diagnosis to classify our case and control animals. This could lead to some misclassification of disease and a loss of power. Since we were able to find numerous significant DMRs, this is likely not impacting us too greatly but should be recognized. Thirdly, due to limitation in the number of dams with transition disease we could find, we did not sample enough calves to follow their specific performance outcomes. We can only apply what we learned from the whole-herd retrospective records analysis and the known functions of DMGs to predict how the DMR identified between TD and NoTD calves may affect future performance.
- A retrospective records analysis of two Cornell University Farms showed dam parity had a larger effect on more aspects of offspring performance than dam disease status and that there was no significant difference in offspring performance between multiparous dams who did or did not have recorded disease in the lactation preceding the daughter’s birth.
- Potential DNA methylation risk loci and changes associated with ketosis were identified in primiparous heifers. One very promising candidate gene for this process is GRINA.
- Potential DNA methylation loci for inheritance of dam disease status were identified in calves at birth born to dams with and without a history of metritis. One very promising candidate gene for this process is CEBPB.
- This project has aided in the education of many undergraduate students, results have been disseminated to research and extension communities, and the project has aided in the professional development of the graduate student project director.
Education & outreach activities and participation summary
Participation summary:
Proposed Outreach Activities:
The main deliverable of this project is to improve our knowledge of the inheritance of DNA methylation and its relationship to health and performance in dairy cattle. This knowledge will help refine genetic selection models for dairy cattle by explaining some of the phenotypic variation due to the environment which is not captured in current prediction models. This will benefit farmers in the future by improving the accuracy of selection as we understand how epigenetics effect phenotypic variation. Within the timeline of this project, dissemination of results to the agricultural and scientific community is more realistic. I will assist in dissemination of results through Dr. Huson’s annual station reports for multistate committee SCC084: Selection and mating strategies to improve dairy cattle performance, efficiency, and longevity, a committee of academic and government researchers who work with industry and advise the Council on Dairy Cattle Breeding of new opportunities to improve genetic selection. Additionally, research results are commonly shared at PRO-DAIRY webinars or events given by Dr. Huson including The Genetic Management of Heifer Reproduction and Implementing Practical Genetics for the Commercial Dairy. These are annual events and webinar series attended by producers, extension agents, and industry representatives. As a way of reaching current undergraduate and graduate students, results of this project will be included in Dr. Huson’s undergraduate Applied Dairy Cattle Genetics course, her annual lecture on large animal genetics to Cornell’s College of Veterinary Medicine students, and my presentations at Animal Science Graduate Student seminar. Publications of this preliminary project may be possible and multiple publications from the future larger study will be submitted to peer-reviewed journals to disseminate results to the broader scientific community. I also plan on attending and can present results at conferences including American Dairy Science Association, American Animal Science Association, International Society for Animal Genetics, and Plant and Animal Genome Conference as applicable.
Outreach Progress Reports:
Outreach Progress (January 2023):
In October 2022, I attended the annual meeting of the multistate research coordinating committee SCC84: Selection and mating strategies to improve dairy cattle performance, efficiency, and longevity at Michigan State University. I presented a brief description of the study design for this project as part of our institution’s station report. I also presented a more detailed description of the study design, sample collection methodology, and background research on the topic of epigenetic inheritance in the Graduate Student Research Updates Seminar of the Animal Science Department at Cornell.
Outreach Progress (January 2024 Report):
In November 2023, I attended the annual meeting of the multistate research coordinating committee SCC84: Selection and mating strategies to improve dairy cattle performance, efficiency, and longevity hosted at our university. I presented a description of the changes to the study design for this project as part of our institution’s station report.
Final Outreach and Education Activity Summary:
Final Outreach Summary:
Since the beginning of this project, I have presented at two multistate research coordinating committee meetings, three Animal Science department graduate seminars, and 1 meeting for Cornell Cooperative Extension Dairy Educators. In January 2025, I will be presenting a poster focused on the DNAm changes identified in calves due to dam disease status at the Plant and Animal Genome Conference. In the summer of 2025, I plan to present at the American Dairy Science Association Annual Meeting about objectives 2 and 4. I am in the process of preparing two journal publications for this project.
Final Education Summary:
Throughout the lifetime of this project, I have trained 12 undergraduate students and 3 graduate students in cattle handling and blood draw techniques who had little to no prior large animal handling experience. I have mentored 5 undergraduate students in the wet lab teaching them DNA isolation techniques. Most recently, I have closely mentored an undergraduate student through most of the analysis for objective 1 of this project and she will be presenting a poster at the Plant and Animal Genome Conference in January 2025. She will also be writing a senior honors thesis on her portion of the analysis, and I will continue to mentor her through that process.
Project Outcomes
This project has improved our understanding of the lasting effects of disease in dairy cattle which has economic, sustainability and welfare implications. Knowing the lasting and intergenerational effects of disease provides additional incentive for disease prevention. This in turn improves animal welfare, lowers costs of production, and improves efficiency of production. In the future, our project and others like it could improve genetic selection practices in dairy cattle allowing us to select for healthier, more sustainable animals.
I plan to continue building on the knowledge gained from this project. My advisor and I have both applied for grants to support this. We hope to investigate other diseases and gain a better understanding of how much DNA methylation changes throughout development in dairy cattle. We believe there is a lot more to learn about how epigenetic information could improve the effectiveness of genetic selection in dairy cattle.