Final report for GS22-256
Project Information
In the coming decades, the demands on global agriculture are expected to escalate from the increase in population and other challenges. The rapid decrease in arable land due to urban expansion and land degradation, and the adverse effects of climate change aggravate worldwide food shortages. Climate change imposes severe crop production limitations, especially for cool-season crops like lettuce in the Southern States. Lettuce quality is extremely susceptible to high temperatures, which restrict their growing areas to only California and Arizona in the U.S. This limitation poses a high risk to the lettuce supply chain as climate change continues to threaten lettuce production. By providing strategies to improve heat tolerance in lettuce, growers can expand lettuce growing seasons and areas, which will alleviate the burden and risk of its supply chain from two western states. Development of heat-tolerant lettuce is highly hampered by the unavailability of ideal breeding materials; therefore we propose the following strategies to enhance sustainable production of lettuce under the changing demographic and climatic situations: 1) suppress bolting under high temperature with foliar delivery of small interfering RNAs (siRNAs) particles, which is a NON-GMO approach; 2) develop heat-tolerant lettuce variety through consumer-friendly biotechnology. The outcome may increase lettuce production to provide more people with higher quality lettuce, reduce greenhouse gas, and reduce water consumption.
Objective 1. Evaluate foliar delivery of siRNA particles for suppressing bolting under high temperatures
RNA interference (RNAi) is a commonly used biotechnology tool to inactivate gene function by degrading their transcripts, thus leading to an expected phenotype. Direct foliar application of naked double-stranded RNA (dsRNA) may efficiently penetrate the plant cells and activate RNAi. A recent report indicated a substantial increase in the effectiveness of gene silencing when spraying plants directly with methylated dsRNA-derived small interfering RNA (siRNA) [6]. The external application of siRNA on the plant’s surface is short-lived and temporarily changes the plant’s internal mechanisms for protection against specific stimuli within a certain period. Successful application of the siRNA system may extend lettuce production time and increase productivity in many southern states. Thus, we will be evaluating a delivery platform utilizing siRNAs methylated at 3’ ends for systemic silencing of miR172 in lettuce to prevent bolting. It is expected that in vitro-synthesized siRNAs to be rapid and reproducible resulting in stable particles that can be applied via canopy spraying in ambient conditions. The miR172-siRNA will be sprayed on Salinas lettuce leaves grown in the field using a hand-held sprayer and bolting time will be recorded. Additionally, lettuce yield will also be measured to determine the effects of miR172-siRNA on lettuce production.
Objective 2. Develop heat tolerant lettuce through consumer-friendly CRISPR genome editing
We will utilize CRISPR/Cas9 genome editing technology to induce double-stranded breaks and create knockout mutations within the miR172 gene which disrupt their functions. Gene-edited lettuce plants will be generated from genetic transformation and plant tissue culture regeneration. The optimized genome editing system in our lab has gene editing efficiency of up to 40 percent with 40-50 transformed plants in the first generation [10]. Thus, it is expected that lines carrying a mutation in miR172 will be identified in T0 plants through sequencing. Segregated homozygous gene-edited lines without Cas9 will be grown in the greenhouse and the seeds will be collected for high-temperature treatments. We expected that the miR172 lettuce mutant will display increased tolerance to high temperature when compared to the wild type. MiR172 mutant and wild-type Salinas seedlings will be subjected to high day/night temperature and measurements such as ion leakage, chlorophyll fluorescence (Fv/Fm), and reactive oxygen species (ROS) will be analyzed to determine heat tolerance between the mutants and the wild type. Additionally, fresh/dry weight and bolting time will also be recorded and compared between the gene-edited plants and wild-type Salinas. Lines that showed high potential for heat tolerance will be propagated for future characterization and field trials.
Research
Experimental design 1.1. In vitro-synthesis of small interfering RNAs (siRNAs)
Arabidopsis miR172 was used to BLAST against the lettuce (Lactuca Sativa) genome to identify the non-coding sequence in lettuce. The identified sequence has been tested successfully in silencing miR172 using the short tandem target mimic (STTM) method in lettuce. For the synthesis of miR172-siRNA, the conserved sequence will be used and sense and antisense stranded will be synthesized separately with perfectly complementary double strands. The miR172-encoding single-stranded RNA (ssRNAs) oligonucleotides will be in-vitro synthesized, methylated, and HPLC purified by GENEWIZ (New Jersey, USA). To form the siRNA duplexes, equal volumes of 100µM miR172-ssRNAs will be combined and annealed at 90 °C for 1 min and cooled to room temperature. The final concentration of annealed oligonucleotides miR172-siRNA will be diluted to 50µM and 100µM before canopy spraying on lettuce plants.
Experimental design 1.2. Foliar application of siRNAs
Salinas lettuce seeds are stored at the Mid-Florida Research and Education Center (MREC). Seeds will be placed on a moistened filter paper and germinated seeds will be planted in the open field located at 28.6° N, 81.5° W. Seedlings preparation and field planting will occur in August and the plants will be ready for spraying in October when the warm temperature can still influence early bolting. In October, two-month-old lettuce will be sprayed with A) No siRNAs; B) 50µM of miR172-siRNA; and C) 100µM of miR172-siRNA. 16 lettuce plants will be randomly selected for each treatment as one replication, and a total of three replications will be applied to each treatment. The experiment will be repeated in January to determine the differences in treatments for two consecutive field trials. Thus, the plants will be sprayed in March with the same treatments. Two weeks and three weeks after siRNAs spraying, lettuce leaf samples will be collected and gene expression of miR172 will be measured using quantitative real-time PCR (qPCR). Lettuce bolting date and fresh/dry weight will be recorded and compared between treatments and between growing seasons. Data will be analyzed using R Studio. A one-way ANOVA followed by Tukey’s HSD will be used to determine whether the bolting date varied significantly between treatments.
Experimental design 2.1. CRISPR/Cas9 design and lettuce plant transformation
As the non-coding sequence of miR172 has previously been identified in lettuce, both CRISPR-P and CRISPOR will be utilized to select two sgRNAs using SpCas9 with Protospacer Adjacent Motif (PAM) “NGG” and the sgRNAs with high rank by two web-tools will be selected and cloned into the SpCas9 expression vector. For plant transformation, Salinas WT seeds will be sterilized with 1.3% hypochlorite, washed with sterilized water, and germinated on MS media. Cotyledons from 5-7 day-old lettuce seedlings will be excised and infected with Agrobacterium tumefaciens EHA105 (with OD600 of 1.5) harboring Cas9-miR172 for 15 mins. The infected cotyledons will be transferred onto the Co-cultivation medium (Murashige & Skoog Basal medium (MS), pH=5.8, Sucrose=3%) for 3 days in the dark at 25˚ C. The infected cotyledons will be further maintained on a callus induction medium for 1-3 months (MS, pH=5.8, Sucrose=3%, BAP=3mg/L, NAA=1mg/L) and any regenerated shoots will be excised, and rooted in soil. Regenerated shoots will be sent for sequencing to identify the gene-edited lines and those lines will be grown in the greenhouse for seeds collection.
Experimental design 2.2. High-temperature treatment of miR172 gene-edited plants vs. wild type
Seeds of gene-edited lines miR172 and Salinas WT will be sterilized with 1.3% hypochlorite, washed with sterilized water, and germinated on MS media. For seedlings heat stress treatments, germinated seedlings will be transferred to Murashige & Skoog Basal medium and maintained at 14 h of light and 10 h dark at optimum day/night temperature (20 °C/15 °C). Eight-day-old seedlings will be subjected to high day/night temperature (37 °C/25 °C) for 24 h and 5 days for short duration high-temperature stress (SDS) treatment and long duration high-temperature stress (LDS) treatment, respectively. Additionally, high temperature-treated plants for 4 days will be shifted back to the optimum temperature for 24 h to get the recovery samples (REC). On the 13th day after germination, 16 shoot tissues samples from each gene-edited line or WT with different treatments (control, SDS, LDS, and REC) will be harvested for RNA extraction as one biological replicate with a total of three biological replicates. Heat-related genes will be measured to determine how the mutation of miR172 affects heat-responsive molecular mechanisms. Physiological measurements such as ion leakage, chlorophyll fluorescence (Fv/Fm), and reactive oxygen species (ROS) will also be analyzed to examine whether the gene-edited lines have higher heat tolerance than the WT. Additionally, fresh weight, dry weight, and bolting time will be measured for a set of at least 16 T2 plants from the gene-edited lines and Salinas WT to determine how the knockout mutation of the miR172 gene affects plant growth and development.
SARE Graduate Student Grant Report
Objective 1. Evaluate foliar delivery of siRNA particles for suppressing bolting under high temperature.
Experiment 1: Testing tobacco rattle virus plasmids for virus induced gene silencing in lettuce through Agrobacterium spraying.
Method: Two sets of tobacco rattle virus RNA plasmids were ordered from Addgene:
- TRV1: https://www.addgene.org/148968/
- TRV2: https://www.addgene.org/148969/
- pLX-TRV1: https://www.addgene.org/180515/
- pLX-TRV2: https://www.addgene.org/180516/
To test the efficiency of the double transformation system (TRV1+TRV2), eGFP was first PCR amplified from plasmid pgwb405 using primers eGFP-F-xbaI (aatctagaATGGTGAGCAAGGGC) and eGFP-R-SacI (atgagctcTTACTTGTACAGCTCGTCC). The PCR product was purified, then digested with XbaI and SacI before ligating to TRV2 and pLX-TRV2 to make TRV2-eGFP and pLX-TRV2-eGFP. The plasmids were transformed into Agrobacterium strain EHA105. It is expected that high GFP fluorescence would indicate the effectiveness of the plasmids and Agrobacterium spraying.
Lettuce (Lactuca Sativa) seeds were germinated on moist filter paper and transferred to 4-inch pots after germination. Four weeks old lettuce was sprayed using a handheld bottle with EHA105 containing TRV1 and TRV2-eGFP combined as the first set and pLX-TRV1 and pLX-TRV2-eGFP as the second set. The plants were grown in a controlled environment with temperatures between 22℃ and 25℃ under white, fluorescent light. GFP fluorescence was checked every day after spraying.
Result: No GFP was observed from Day 1-20 after spraying (Figure 1).
Figure 1: White light and GFP images of two sets of tobacco virus plasmid. No GFP fluorescence was observed.
Summary: Due to the low efficacy of the tobacco virus plasmids in lettuce, it was determined that another method should be tested to evaluate foliar delivery of RNA for suppressing bolting of lettuce under high temperature. Studies have shown that plants can uptake double stranded RNA that mimic or inhibit microRNA since the mature sequence of the microRNAs are short and can travel throughout the plant. As a result, the expression level of the microRNA will be increased or reduced and subsequently affecting the target genes. Thus, experiment 2 was carried out using synthetic matured microRNAs 156 and 172 sequence without any plasmid and Agrobacterium transformation.
Experiment 2: Testing double stranded RNA to increase miR156 expression and inhibit miR172 to prevent bolting in lettuce.
Method: Arabidopsis matured miR156C and miR172 RNA sequences were synthesized with chemical modification to the sequences to enhance their stability.
-AtmiR156C mimic single stranded RNA with 2’-O-methyl RNA (2’OMe) was synthesized and annealed at 95℃: RNA mature strand: 5’-UGACAGAAGAGAGUGAGCA[2OMeC]-3’; RNA complementary strand: 5’-GCUCACUGCUCUAUCUGUCAG[2OMeA]-3’
-AtmiR172 double stranded RNA was synthesized with 2’OMe and ZEN modification: RNA mature sequence: AUGCAGCAUCAUCAAGAUUCU; sequence with chemical modification: mU/ZEN/mAmUmGmCmAmGmCmAmUmCmAmUmCmAmAmGmAmUmUmCmU/3ZEN/
In addition to testing bare double stranded RNA, carbon nanoparticles were mixed with the RNAs to evaluate the effectiveness of carbon nanoparticles in delivering the microRNAs into the plant. Nanoparticles have been shown to act as a carrier to deliver particles such as nutrients through cell wall thus promoting plant growth. Lettuce seeds were germinated on moist filter paper and potted in 8-inch pots. The plants were grown in a greenhouse located at the Mid-Florida Research and Education Center in Apopka, Florida from January to April 2023. Four weeks old lettuce seedlings were treated with water as the control, 250mg/L of carbon nanoparticles (CNP), 0.5µM miR156, 0.5µM miR156 with 250mg/L carbon nanoparticles, 0.5µM miR172, and 0.5µM miR172 with 250mg/L carbon nanoparticles by pipetting 100uL of each treatment directly on top of the apical meristem two times a week until flowering. Six plants were used for one replication for each treatment and the experiment was repeated three times for a total of 18 plants per treatment.
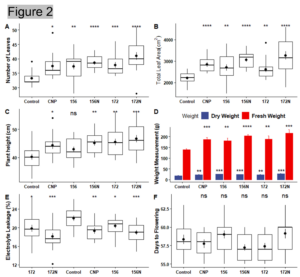
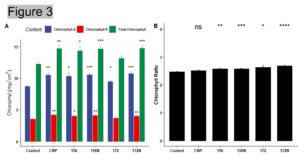
Result: Measurements such as number of leaves, leaf area, plant height, fresh/dry weight, and days to flowering were recorded and analyzed. Electrolyte leakage and chlorophyll content were also measured. All the treatments showed a higher number of leaves and total leaf area when compared to the water control (Figure 2 A and B). Similarly, the treated lettuces were taller and weighed more than the water control (Figure 2 C and D). Overexpression of miR156 and silencing of miR172 are well known to affect flowering time. However, the flowering date did not differ between the treatments (Figure 2 F). While there was no significant difference in flowering date, higher chlorophyll content and lower electrolyte leakage in the treated plants suggested that the plants were able to tolerate environmental stress with either carbon nanoparticles and/or microRNAs (Figure 2 E and Figure 3). Representative images of the treated plants can be seen in Figure 4.
Figure 2. Morphological and Physiological measurements. (A) Number of Leaves. (B) Total Leaf Area. (C) Plant height. (D) Fresh and Dry Weight. (E) Electrolyte Leakage (F) Days to Flowering.
Figure 3. Chlorophyll measurements of treated lettuce (A) Chlorophyll a,b and total chlorophyll (B) Chlorophyll a/b ratio
Figure 4. Representative images of water control, carbon nanoparticles (CNP) and microRNA treated plants in lettuce.
Summary: It was evident that the addition of either carbon nanoparticles, microRNA, or both promoted plant growth resulting in bigger lettuce plants that can tolerate environmental stress. How each treatment affected the expression level of miR156 and miR172 are currently being investigated through qPCR. From the promising results, it was determined that the experiment will be repeated with higher concentration of RNA to prevent bolting in lettuce. Additionally, another plant species will also be tested to evaluate the effectiveness of the treatments across different plant species.
Objective 2. Develop heat tolerant lettuce through consumer friendly CRISPR genome editing.
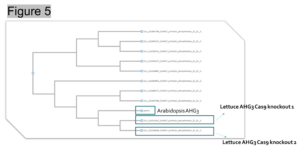
Method: The original objective was to use CRISPR/Cas9 to knockout miR172 in lettuce to prevent bolting. However, considering the involvement of miR172 in different plant growth and development processes, miR172 mutants may have severe deformity. After evaluating the pathway of miR172, a candidate gene ABA-hypersensitive germination3 (AHG3) was selected for gene synthesis and CRISPR/Cas9 knockout. By blasting against Arabidopsis AHG3, it was found that there were two homologs of AHG3 in lettuce (Figure 5). To ensure the complete knockout of AHG3 in lettuce, sgRNAs were designed to target both homologs so that mutation can be created in both locations (Figure 6).
Figure 5. Two AHG3 lettuce homologs were found closely related to Arabidopsis AHG3.
Figure 6. Sequence including sgRNAs for AHG3 for gene synthesis to use with the CRISPR/Cas9 system.
Summary: A new candidate gene, AHG3 was selected as the target for CRISPR/Cas9 knockout to prevent bolting in lettuce. DNA sequence was synthesized by GeneWiz. Vector cloning and plant transformation will be carried out in the next six months before regenerated plants can be sent for sequencing to confirm knockout mutation of AHG3.
Objective 2. Develop heat tolerant lettuce through consumer friendly CRISPR genome editing.
Plant Transformation Materials and Methods: Seeds of lettuce (L. sativa cv. Salinas) were obtained from UC-Davis and grown at the Mid-Florida Research and Education Center in Apopka, FL for seeds. Newly collected seeds were stored in sealed containers at 15 °C until used. Seeds were surface-sterilized with 70% alcohol for 2 min, followed by 1.3% hypochlorite solution for 10 min; seeds were washed with sterile double-distilled water 5−8 times prior to germinating them on MS (Murashige and Skoog) media with 3% sucrose and 0.8% agar at pH 5.8. Genetic transformation and regeneration were modified from the protocol developed by Michelmore and Marsh. In brief, 12−14 days after germination, cotyledons were cut and dipped into Agrobacterium strain EHA105 harboring the intended plasmid for 10 min; the infected cotyledon was subsequently cultured on co-cultivation media (MS + 3% sucrose) in the dark for 2 days, before transferring to shooting media (MS + 2 mg/L 6-benzylaminopurine + 0.1 mg/L NAA + 100 mg/L Kanamycin and Timentin).
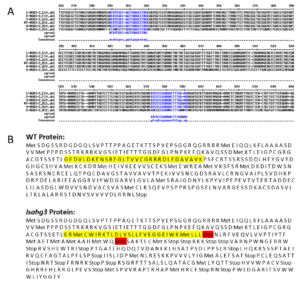
Gene Edited Plants Confirmation: Rooting of newly regenerated shoots was carried out on a half-strength MS medium containing 100 mg/L kanamycin but with no supplemental hormones. GFP fluorescence was detected with a fluorescent stereomicroscope (Leica, German) and images were captured using Nikon D800 digital camera attached to the microscope. After in vitro rooting of regenerated shoots, they were potted in soil and kept at constant room temperature with 16 h supplemented incandescent light for two weeks before repotting in 1-gallon pots to grow in the greenhouse. T1 seeds were germinated on filter paper moistened with distilled water and fully expanded cotyledons were used for DNA extraction using Edwards’s Buffer. PCR products of primers flanking the targeted AHG3 genes were sent to Genewiz, Inc (South Plainfield, NJ, USA) for Sanger sequencing. Sequencing results were aligned using EMBL-EBI and Synthego Performance Analysis (ICE Analysis. 2019. v2.0. Synthego) was used for editing analysis. Sequencing alignment between five lsahg3 mutants and WT showed no gene editing occurred at the sgRNA1 location, and one base pair deletion at sgRNA2 location in the lsahg3 mutant (Figure 1 A). Consequently, the deletion at sgRNA2 leads to alteration in protein translation and premature stop codon rendering the protein nonfunctional (Figure 1 B).
Figure 1: Sequencing alignment between the WT and lsahg3 mutant. (A) DNA sequence alignment of WT and mutants. Blue indicates misalignment, dash indicates bp deletion. (B) Protein alignment of WT and mutant, yellow highlighted the start of protein misalignment and Stop codon is highlighted in Red.
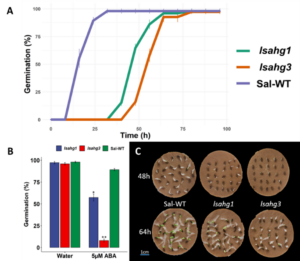
Seed Germination Phenotype: In a seed germination test, the previously created lsahg1 mutant and newly created lsahg3 mutant were evaluated against Sal-WT lettuce to assess the functional consequences of these mutations on seed germination dynamics and abscisic acid (ABA) sensitivity (Figure 2 A). The germination rate revealed a marked delayed germination in the mutant lines, while the WT achieving 100% of germination within 25 hours, lsahg1 followed closely by lsahg3 took almost 75 hours post imbibition for 100% germination rate. Interestingly, the differential response in the presence of 5µM ABA—a known inhibitor of germination—was pronounced, as both lsahg1 and lsahg3 displayed a significant susceptible to ABA-mediated suppression of germination, suggesting an altered ABA signaling pathway (Figure 2 B). These findings, visually corroborated by germination phenotypes across temporal checkpoints at 48 and 64 hours, elucidate the roles of AHG1 and AHG3 in modulating the timing and sensitivity of germination through ABA-dependent mechanisms (Figure 2 C).
Figure 2. Germination assays of lettuce seeds comparing CRISPR/Cas9 knockout mutants lsahg1 and lsahg3 with Sal-WT (wild type). (A) Time-course of seed germination rates between Sal-WT and the mutants. (B) Germination percentages at a fixed time point under control (water) and 5µM ABA treatment, highlighting the ABA sensitivity of the mutant lines. (C) Representative image of the physical germination states at 48 and 64 hours, with the mutants exhibiting delayed germination compared to Sal-WT.
Challenges in Growing Lettuces in Florida and Obtaining the Mutant Lines: Salinas is a head lettuce and the high humidity levels typical of Florida promote rotting in head lettuce. Figure 3 A highlights rotting areas, while the close-up view reveals darkened, decayed tissue. This rotting is likely caused by excessive moisture creating an environment conductive to fungal and bacterial growth, leading to tissue decay. Thus, the head of Salinas must be continuously open to prevent moisture and rotting. A major pest of lettuce in Florida was observed to be whiteflies. Figure 3 B shows whitefly infestations, identified by the small, white insects on the leaves, while the close-up view clearly displays whiteflies on the leaf surface. Whiteflies not only directly damage plants by feeding on their sap but also act as vectors for plant viruses, compounding the threat they pose to plant health. Figure 3 C illustrates pathogen infections in lettuce, with visible signs of leaf discoloration and damage. The top image shows general leaf damage, while the close-up view highlights spots and lesions indicative of disease. These symptoms suggest diseases such as downy mildew, which thrive in Florida's warm and humid environment. Even with constant spraying and greenhouse management, the humidity and heat in Florida increases pests and pathogens activities in lettuce which lead to poor plant development and ultimately plant death.
In addition to the problems mentioned above, lettuce also suffered poor pollination and seed development during summer in Florida. As a result, it was difficult to maintain healthy plants and minimal seeds were collected from the lettuce plants in the summer. Thus, all the Salinas WT and mutant lines can only be grown from September to May in the greenhouses. Due to the challenges in growing lettuce, flowering phenotype data collection was delayed and only seed germination phenotype was recorded near the end of the grant proposal.
Figure 3. Representative images of challenges in growing lettuce in Florida. (A) Humidity in Florida increases rotting in head lettuce. (B) Insects such as white flies in lettuce. (C) Pathogens in lettuce.
Educational & Outreach Activities
Participation summary:
The tour was given to the Sweetwater Oaks Garden club, where members of the club who supported the University’s students financially were able to learn about the student’s project and their importance. The members were excited about the potential of the project and gained insight on how the experiments were conducted.
Undergraduate students were trained in different lab skills including morphological and physiological measurements of the treated plants. They developed an understanding of experimental design and data collection.
Project Outcomes
The results will contribute significantly to the social benefits of farmers by producing a new variety of lettuce that can tolerate heat stress so that farmers can expand their lettuce growing seasons and have higher economic returns. The microRNA strategy can reduce plant stress without toxic chemicals resulting in better lettuce quality. These strategies will also expand lettuce growing areas to southern states giving farmers more options and reduce carbon emissions from transporting lettuce across the state.
During this project, the student gained experimental design skills and knowledge involving the flowering time pathway of lettuce including microRNA 156 and 172. From the promising preliminary results, the student and advisor believe that it’s highly possible to alleviate heat stress in lettuce using alternative strategies such as gene editing and microRNA delivery for a more sustainable lettuce production.