Progress report for GS23-278
Project Information
Recently, strawberry growers in Florida have incurred severe yield losses due to unprecedented outbreaks of leaf spot and fruit rot caused by a new species of Neopestalotiopsis. The failure of most fungicides registered for strawberry and the susceptibility of current cultivars demand breeding for improved genetic resistance. Fortunately, the University of Florida strawberry breeding program has identified a source of resistance to Neopestalotiopsis sp., an Israeli cultivar named ‘Yasmin’, and confirmed that resistance is heritable in breeding populations derived from ‘Yasmin’. We propose screening Yasmin-derived populations for resistance in inoculated field trials. Analysis of field resistance and genetic data via association mapping will approximate the locations of genes underlying resistance. Mining these locations for DNA diagnostic information in future work would enable breeders to effectively select for Neopestalotiopsis sp. resistance in their breeding populations. This research will support ongoing efforts to efficiently and more rapidly deliver cultivars with improved Neopestalotiopsis sp. resistance.
The overall aim of this project is to uncover genetic markers conferring resistance to Neopestalotiopsis sp. as targets for breeding resistant cultivars.
Objective 1: Evaluate Yasmin-derived populations for Neopestalotiopsis sp. resistance in replicated, inoculated field trials.
Objective 2: Identify genetic regions increasing resistance to Neopestalotiopsis sp. and characterize their phenotypic effects in Yasmin-derived populations.
Objective 3: Select individuals with superior resistance and other breeding-relevant traits for further backcrossing into elite backgrounds.
Cooperators
- (Researcher)
Research
- Sixteen full-sib families, generated from biparental crosses between Yasmin or progeny of Yasmin and 12 UF advanced selections with excellent yield and fruit quality, are retained as mother plants in the UF summer breeding nursery near Malin, Oregon. To develop large field populations, full-sib families, parents, and susceptible checks were clonally propagated via runners in the nursery. Five clonal replicates of each seedling were then harvested during mid-September and transported to the UF/IFAS Gulf Coast Research and Education Center (GCREC) in Wimauma, Florida where they were stored at 3.5°C until planting. Transplanting took place on 19 Oct. 2023 in a randomized complete block design with one runner plant in each of five blocks at GCREC. Each block consisted of a single raised bed (350 feet long and 3 feet wide) covered with plastic mulch. Plants were established with overhead irrigation during daylight hours for 10 days after planting, followed by drip irrigation for the rest of the growing reason. Once the seedlings were established, they were inoculated with an atomizer by spraying each plant with 2 ml of spore suspension (2 x 104 spores/ml). Spore suspensions were prepared from 7- to 10-day-old colonies of four Neopestalotiopsis sp. isolates revived from the culture collection at GCREC on potato dextrose agar under continuous fluorescent lighting at room temperature. After inoculation, plants were overhead irrigated for 10 days to encourage spore dispersal. Disease severity was assessed at four timepoints post inoculation based on the following 0-7 rating scale: 0, no symptoms; 1 = 1 to 10%, 2 = 11 to 25%, 3 = 26 to 50%, 4 = 51 to 80%, 5 = 81-100% of overall leaf area (per plant) covered by lesions, and 6 = dead. Phenotyping was initiated with the onset of symptoms on check cultivars and continued on a weekly or biweekly basis for four weeks post inoculation. A single area under disease progress stairs (AUDPS) value per individual was calculated by considering the severity score of all clonal replicates at all timepoints.
- To detect DNA variants controlling Neopestalotiopsis sp. resistance, we will analyze all segregating populations via genome-wide association mapping (GWAS). A GWAS requires two data sources: DNA sequence information and seedling phenotypes. To obtain the first, DNA will be extracted from all seedlings according to Mangandi et al. (2017) and submitted for genotyping using the FanaSNP array through Affymetrix. The FanaSNP array screens each seedling’s DNA with 50,000 single-nucleotide polymorphisms (SNPs) commonly segregating in cultivated populations. SNP genotypes of each seedling will then be reported for the 50,000 examined locations across the strawberry genome. Coupled with phenotypic data collected from our seedling trial, SNP genotypes will be analyzed via GWAS, leading to the discovery of genetic variants that associate with Yasmin-derived resistance. The genetic effects and heritability of associated markers will then be estimated in statistical models to inform the next steps of breeding for Neopestalotiopsis sp. resistance.
- Distributions of disease severity values were plotted for the progeny populations and compared against the susceptible and resistant parents’ mean values. Individuals with superior Neopestalotiopsis sp. resistance and adequate flower production were selected as donors of resistance in future crosses with proven, elite breeding parents, to recover desirable yield and fruit quality traits in successive generations.
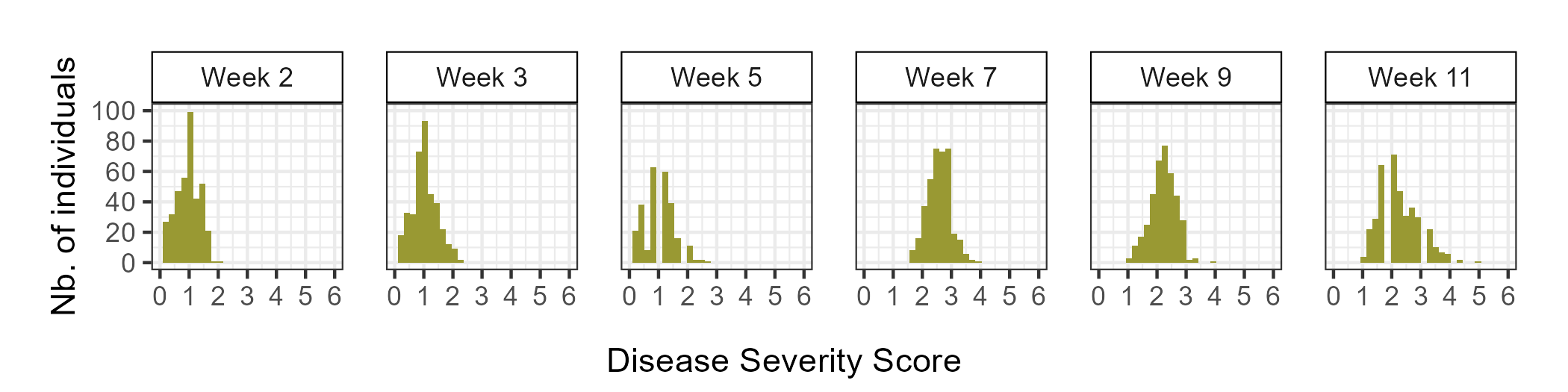
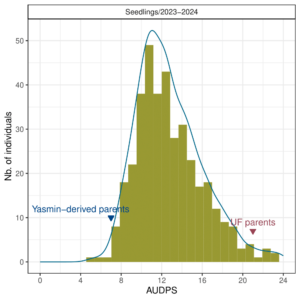
 During the 2023-2024 strawberry production season, a total of 382 individuals were screened for resistance to Neopestalotiopsis sp. Individuals were categorized as either resistant (0 ≤ ȳ ≤ 2) or susceptible (2 < ȳ ≤ 6), with ȳ representing the mean of disease severity scores among replicates. Plants with a ȳ of 0 showed no symptoms, while those with a ȳ of 6 were dead. Out of the 382 individuals evaluated, 162 (42%) displayed mild symptoms (0 ≤ ȳ ≤ 2) and were classified as resistant, while 220 (58%) exhibited moderate to severe symptoms (2 < ȳ ≤ 6) and were classified as susceptible (see Figure 1). The mean values for the Area Under the Disease Progress Curve (AUDPS), a metric that tracks disease symptom progression over time, revealed continuous distributions, skewed towards higher values indicative of increased disease severity due to wet conditions in Central Florida during the 2023-2024 production season (see Figure 2 and 3).
During the 2023-2024 strawberry production season, a total of 382 individuals were screened for resistance to Neopestalotiopsis sp. Individuals were categorized as either resistant (0 ≤ ȳ ≤ 2) or susceptible (2 < ȳ ≤ 6), with ȳ representing the mean of disease severity scores among replicates. Plants with a ȳ of 0 showed no symptoms, while those with a ȳ of 6 were dead. Out of the 382 individuals evaluated, 162 (42%) displayed mild symptoms (0 ≤ ȳ ≤ 2) and were classified as resistant, while 220 (58%) exhibited moderate to severe symptoms (2 < ȳ ≤ 6) and were classified as susceptible (see Figure 1). The mean values for the Area Under the Disease Progress Curve (AUDPS), a metric that tracks disease symptom progression over time, revealed continuous distributions, skewed towards higher values indicative of increased disease severity due to wet conditions in Central Florida during the 2023-2024 production season (see Figure 2 and 3).
Our finding that a significant percentage (42%) of Yasmin’s progeny populations exhibited resistance suggests that this trait is controlled by one or more moderate- to large-effect resistance loci. The introduction of these putative loci into UF germplasm has apparently expanded the range of observed phenotypes in the lower direction (towards resistance) as observed in the disease severity distribution (Figure 2), where symptomless individuals were observed even 11 weeks post-inoculation with the aggressive pathogen. Meanwhile, UF check cultivars had developed severe leaf and fruit spots, and some were already dead. A forthcoming genome-wide association study will pinpoint the specific loci and genetic markers contributing to resistance against Neopestalotiopsis sp. These loci can then be directly targeted in breeding towards developing resistant cultivars. Furthermore, we have identified promising individuals within Yasmin’s progeny populations that combine superior resistance to Neopestalotiopsis sp. with several other traits of interest to the program, such as high yield and quality fruit, that will immediately be used in crosses as parents.