Progress report for GS23-295
Project Information
Irrigation can be a short-term solution to water scarcity. For many areas of the world, long-term irrigation use can be unsustainable for cotton agriculture. Cotton typically has some drought tolerance; however, the inability to easily phenotype the root system architecture (RSA) has resulted in elite cultivars with reduced growth and plasticity in root systems. To explore cotton’s phenotypic variation of RSA in the germplasm, 660 accessions from around the globe were narrowed down to 200 accessions for RSA phenotyping analysis. Four seed companies each contributed three of their elite cultivars to this research. The accessions were simultaneously grown in the greenhouse and field locations. The greenhouse evaluated baseline RSA under well-watered conditions. The field tested the accessions under drought stress and compared the RSA of seven accessions. The rainfed condition of West Texas during the 2024 summer season allowed sufficient drought stress to affect most of the accessions significantly. A minimum of fifteen accessions had no significant difference between the irrigated and rainfed conditions. Many accessions also had a more robust RSA with higher root length, greater root-to-shoot ratio, and finer average diameter. A pattern of thicker primary root diameter, but finer lateral roots, performed better under drought conditions, along with a higher root length and a higher root-to-shoot ratio, which contributed to performing better under drought conditions. Using the phenotypic data of this study will allow for genetic associations to be completed, and identification of potential genes involved in the RSA will help future research and breeders develop cotton accessions that can integrate the benefits of an increased RSA for drought tolerance.
First Year's Objectives
1) Identify a subset of Upland cotton accessions (175-200) and elite commercial cultivars (15) by selecting from the 600 accessions grown in the greenhouse.
2) a) Evaluate the root system architecture (RSA) of the diverse germplasm using image-based root phenotyping platform under controlled greenhouse conditions. The plants will be grown in the greenhouse for about 2 weeks before scanning their RSA with the scanner and software program. b) Select a subset of the 5 most promising and the 5 least promising of the exotic accessions in addition to the control and elite cultivars for contrast. To evaluate the RSA traits, under well-watered and water-deficit treatments in field conditions, five replications will be under irrigated conditions and the other will be rainfed only.
3) Based on the 1st year’s findings, lines with contrasting RSA will be further explored to identify QTLs or genomic loci using advanced genomic tools (GWAS) and additional field experiments to identify novel genes for optimizing the cotton RSA for enhanced water capture and efficiency.
Second Year's Objectives
Objective 1: Evaluation of Root System Architecture (RSA) under greenhouse conditions
Objective 2: Evaluation of selected accessions under field conditions
Objective 3: Analysis of phenotypes and genotypes through genome-wide association mapping
These objectives are the foundational elements of a continuous research project to understand our long-term goal: identify novel QTL/genes for optimizing cotton RSA for enhanced water capture and use efficiency. Based on the 2nd year’s findings, encompassing morphological, anatomical, and physiological traits, lines with contrasting RSA will be further explored to identify gene/QTLs using advanced genomics tools and additional field experiments with continued support from TSSC and other sources.
Research
In the first year, the objectives were to determine the cotton varieties for this project;660 cotton accessions were ordered from USDA-GRIN. Those were planted in the greenhouse to grow for seed accession. Those were screened using phenotypes and their origins to reduce to 200 accessions to move to the next objective. The elite commercial cultivars grown in West Texas were collected from various seed industry partners, including BASF, Bayer, Armor, Phytogen, and NexGen. These companies selected varieties they were interested in testing for drought tolerance in this project. Some varieties already have some drought tolerance characteristics. The main goal of this objective is to get a diverse set of genetic variation to compare root systems for the best and the worst RSA, as well as to compare water efficiency for sustainable water use. Traits related to yield, disease resistance, and fiber quality will also be considered.
For the second objective, all the selected lines were screened for root morphology at the seedling stage, 2 weeks after planting. They were screened using a visual scoring and high-throughput phenotyping platform, WinRhizo, Inc. WinRHIZO is an image analysis system specifically designed for measuring root morphological traits. We focused on total root length, diameter, and surface area among other characteristics. According to Kadam et al. (2015), this process is done in Dr. Jagadish’s lab, which has extensive experience working with root morphology and anatomy of various crops. These experiments will help us narrow the lines for in-depth analysis under well-watered and water-deficient conditions.
The next phase of objective two is to identify lines with differential response to water deficit conditions regarding an altered RSA. To achieve this goal, we plan on growing a subset of contrasting lines in controlled-environmental greenhouse conditions under well-watered 100% field capacity and water-deficit conditions around 40%-60% field capacity (Kadam et al., 2015). Water deficit stress will be imposed after seedling emergence. Plants under the control treatment will be maintained at 100% field capacity throughout the experiments. After 30 days of stress, plants will be harvested around 50 days after sowing, and traits associated with the RSA, such as shoot biomass, will be measured. Physiology will be collected as detailed in Kadam et al., 2015. The anatomical studies will consider the root diameter, stele diameter, and metaxylem number.
After evaluating RSA and anatomical traits from objective 2, cotton cultivars showing differential or contrasting phenotypes were further evaluated under field conditions. Uprooting cotton plants in the field would have been challenging, so large woven bags filled with field soil were used to simulate the field conditions to overcome this issue. Watering was done manually by adding the recommended amount determined by the University of Georgia.
During the cotton growth, the morphological (shoot) traits were measured. These accessions were grown and intended to be harvested at the boll formation stage (i.e., the peak root biomass accumulation phase). However, a hailstorm destroyed some of the bags, and some were damaged. The time it took to wash these manually was longer than expected. Therefore, this experiment had to be repeated over the winter in the greenhouse with the same field soil and the same bags.
The repeated experiment was done in the greenhouse because of the winter conditions. With the limited space in the greenhouse and the amount of soil needed to transfer to the greenhouse from the field, the number of genotypes was reduced to 12 instead of the original 24. This was intended to have three replications of 12 rather than fewer replications. Watering was done similarly to manually measuring the field with the graduated cylinder. The volume and weight were calculated for the soil density, and the soil was set to fit within the same mass per volume to get the ideal density and compaction.
A comprehensive analysis of diverse germplasm and elite commercial cultivars grown in West Texas will allow us to identify the functional role of root tissue plasticity in adapting to water-deficit stress. It will be interesting to evaluate whether current commercial cultivars, in addition to deep root systems, possess active shallow roots and whether this helps conserve soil moisture during the vegetative and reproductive stages.
This information will help investigate, understand, and optimize root plasticity in upland cotton and translate this knowledge to develop next-generation cotton cultivars with better RSA under water-deficit conditions to improve sustainable agriculture. Commercial producers can apply this research to their ongoing projects to improve the future elite varieties, enabling farmers to reduce their water consumption and save money while increasing their yields.
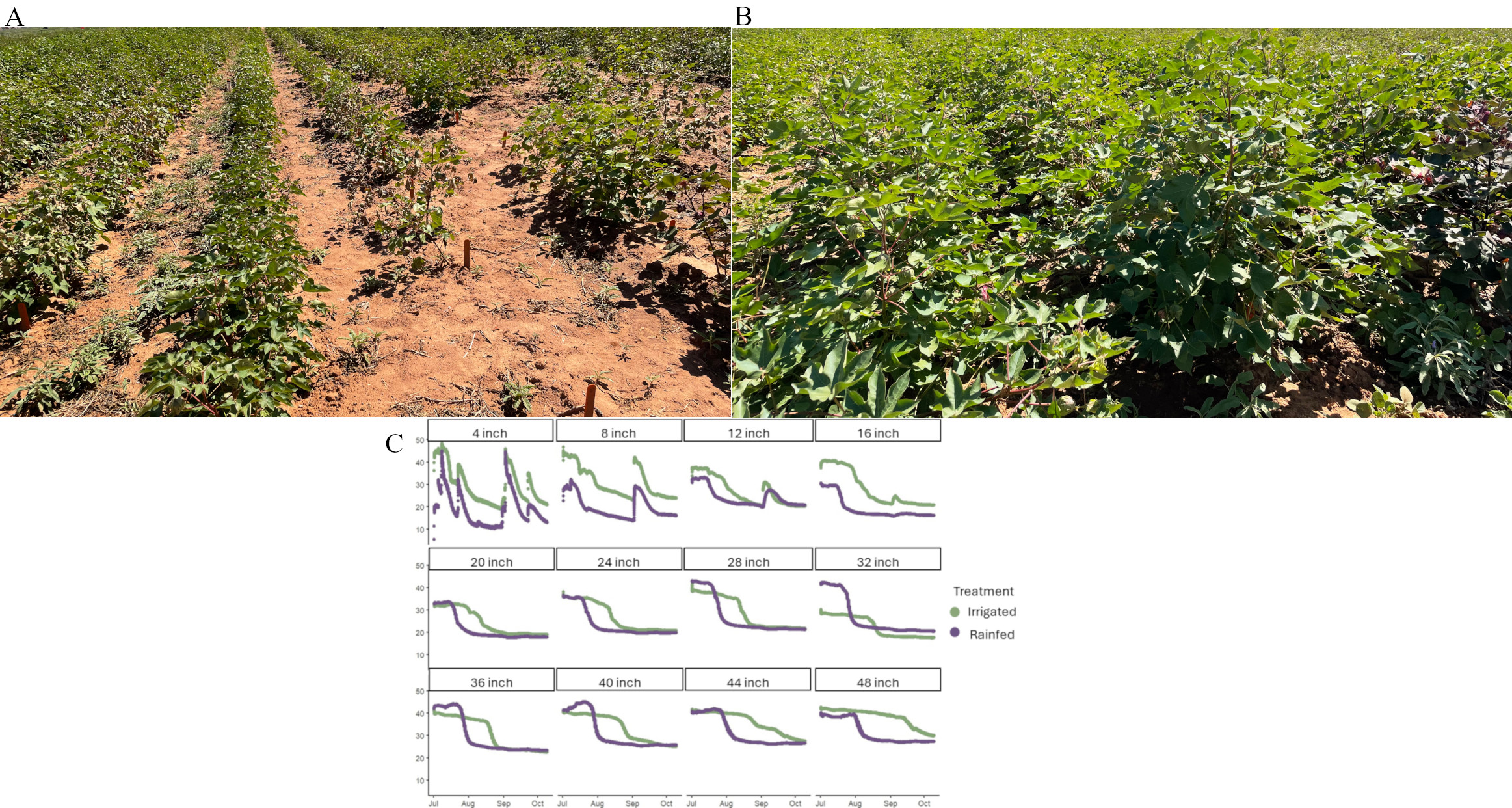
At the end of December 2023, the top 0-12 cm depth of topsoil was moved from the Quaker Research Farm to the Horticulture Greenhouse so field soil could be used for the repeated aboveground experiment to get the root architecture traits for the selected accessions. Accessions were previously selected based on their differences in root phenotypic traits. At the end of the 2023 summer season, many of our replications were destroyed in a hailstorm, and the timing for extraction prevented getting good, replicated data to move forward. The bags were filled with field soil by measuring mass and volume to have as similar a density and compaction as manually allowed (Fig. 1 A&B). 12 Genotypes were used three elite cultivars, TM1, and Coker 312 were used for the reference genotypes, also 3 of the lower and 4 of the higher performing root traits. They were randomly placed for three replications (Fig. 1C).
|
C) |
Row 1 |
Row 2 |
Row 3 |
|
1 |
189 |
141 |
184 |
|
2 |
107 |
70 |
105 |
|
3 |
99 |
132 |
197 |
|
4 |
193 |
66 |
181 |
|
5 |
70 |
105 |
197 |
|
6 |
141 |
66 |
181 |
|
7 |
189 |
184 |
132 |
|
8 |
107 |
99 |
193 |
|
9 |
105 |
181 |
107 |
|
10 |
70 |
197 |
141 |
|
11 |
66 |
184 |
193 |
|
12 |
99 |
132 |
189 |
Figure 1: Bag Set up in the Horticultural Greenhouse with three bags across and 12 bags down. A 5-gallon bucket was used to measure the mass of soil that went into each bag. And the height of the cylinder was measured by marking the inside of the bag. Similar soil density and compaction must be improved to get the same amount of soil mass within a given volume for each bag. B) Representing the three bags across and how the bags look when filled. C) Random design of 3 replications
Each replication was planted two days apart to allow time for extraction for 2 days for each replication. Trying to allow them to be at the same growth stage when extracted. Plants were grown for six weeks and were extracted before they started the flowering stage. Many varieties differ in their flowering stages, and it was intended to see their root traits before allocating resources to the reproductive system. For manually watering the plants, a graduated cylinder was used to give them their recommended inches once per week. Above-ground traits were documented at two growth points before extraction. Plant height and health, number of nodes, level of pubescence, anthocyanin present, and number of leaves are some documented traits (Fig. 2).

When extracting the samples, some difficulties arose with the stickiness of the soil, combined with how delicate the roots were. This made collecting the complete root system intact impossible. Nearly all the roots broke off from the primary root. They were manually collected from each bag until no more roots could be found (Fig. 3). The roots were washed by placing them into a bin of water with a mesh over the top, allowing the fine soil to fall away from the root and keeping the roots moist until time for imaging.

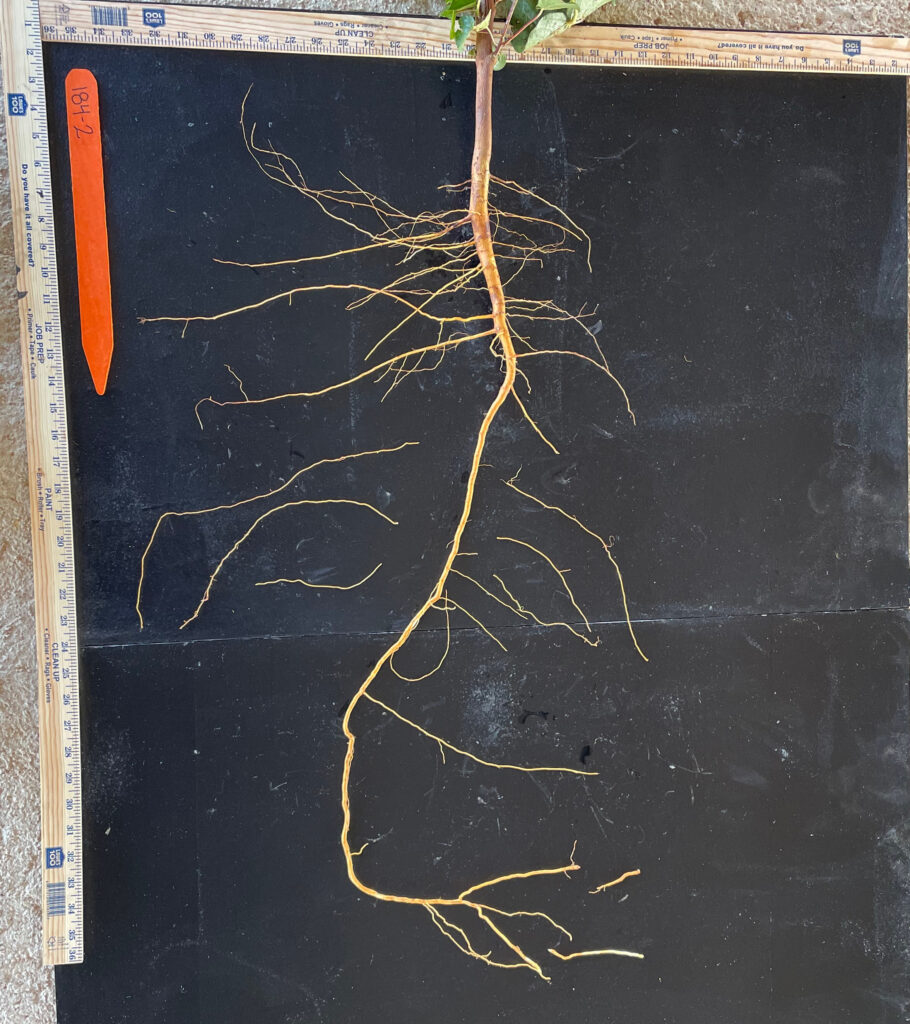
Each root piece was manually placed on a painted black metal sheet (Fig. 4). They were carefully placed so as not to touch each other to obtain the total root length, surface area, and average diameter. Unfortunately, the correct root angle, number of tips and forks, and tap root depth could not be obtained.

These accessions were intended to be placed in this year’s field experiment to see how their performance was under drought conditions; however, attempts to get enough seeds from some of the landrace varieties (105, 107, 099, 141) in time for this year’s field season were unsuccessful. Only a couple of seeds were collected because of their late flowering due to the assumption of photoperiod sensitivity traits. Therefore, accessions 105, 107, 099, and 141 were planted earlier than the previous year, hopeful that more seeds would be collected for future experimentation.
Shortly after removing the tall bags from the greenhouse, another experiment was conducted to collect the RSA phenotypes for the GWAS analysis. Two hundred twelve accessions, 12 elite cultivars, 200 various accessions from obsolete cultivars, landraces, breeding lines, and TM1 and Coker 312, were selected to represent cotton germplasm from around the world and West Texas (Fig. 5). These accessions were selected based on their ability to flower in this area and their similarities and differences in visual phenotypes.

The bags used were 2 gallons filled with the Quaker Research Farm soil. Three replications of each accession were randomly distributed throughout the four tables in the same greenhouse room. Each day, 64 bags were completed. Five seeds were placed in each of the 636 bags. After germination, the weakest seedlings were thinned out, leaving only one plant per bag. This experiment attempted to keep the conditions optimal for well-watered conditions for all accessions because there was not enough room or time for another 636 bags for drought conditions. Each bag was saturated with ~ 600 ml and left to dry for 24 hours to reach field capacity before planting the seeds. Each set of 64 bags was one day offset from the previous 64 bags to allow them to grow the same amount of time and be extracted on the same days after planting (DAP). Within 11 days, all bags were planted (Fig. 6A). Each bag received 200 ml of water on 3, 8, and 15 DAP as the greenhouse temperature would reach 110 degrees during the day. Water was added using a graduated cylinder to ensure the same amount was added to each bag. Plants were extracted on 17 DAP (Fig. 6B). Washing the roots was done by first soaking the bags, allowing them to become overly saturated to reduce the stickiness of the soil to the roots. The bags were cut and ripped away as one held onto the plant's stem. Then, working the way down the tap root, try to loosen as many roots as possible from the soil and gather any roots that broke off. Samples were stored in Ziplock baggies with 50% ethanol and placed into a 4° C room until they could be scanned (Fig. 6C).

Samples were scanned the same way as the previous year's sample with the WinRHIZO scanner and software by placing an acrylic tray with some water to ease the roots' separation. Plastic tweezers were used to pry apart the roots to prevent all bunching up in one section. However, some roots were so prolific that they made separation challenging. Each sample took around 7 minutes to scan, and while one was being scanned, another sample was being prepped and separated.
The same 212 accessions grown in the greenhouse were also planted in the Quaker Research Farm in a 16-row plot with sub-surface drip irrigation that I could manually turn on or off for eight rows at a time (Fig. 7). For the first eight rows, I left the irrigation off for the entire season and only received water from the rain. In the second 8 rows, I supplemented the weeks with no rain with irrigation. Aimed for about 1” of water per week. The irrigated plot received 5 weeks of irrigation more than the rainfed plants.

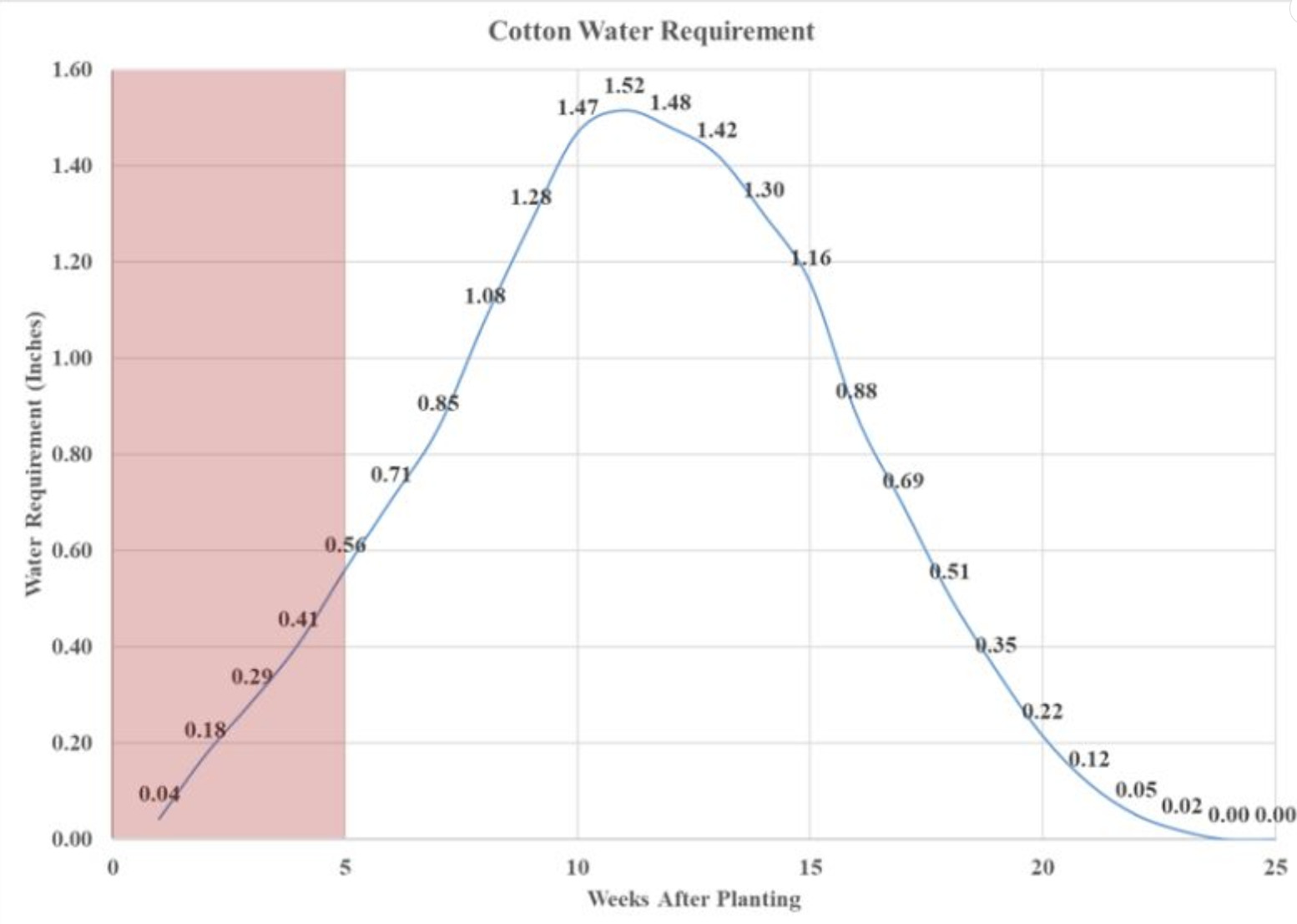
Figure 8 shows the difference in canopy growth between the two plots in September. Irrigation was stopped at the beginning of September. A sensor from AquaSpy was placed in the rainfed and irrigated fields to take measurements until the end of the season. It measured every 15-minute interval with moisture levels, salinity, and temperature. Figure 8 focuses on the moisture levels collected by both sensors. The sensors were placed at the beginning of July; irrigation began four weeks after planting.
By the peaks at 4 inches depth, it is possible to determine when it rained during the months of July through October. There was no rain during the hottest months of the year, from August through September, contributing to the drought stress for the cotton during this time. The difference in water between the treatments through August and September is a clear separation at most depths. Looking at the 8 to 12-inch depth where the drip irrigation emitters are located, there are two steady points in the month of July. But a slow decrease in August, which could suggest that the 1 inch of irrigation was not enough to bring it back up to saturation levels, but most likely helped to prevent the plants from being too stressed during August. A repeated experiment next summer, with time to do physiological measurements, would be helpful to know the stress of the leaves between the two treatments.
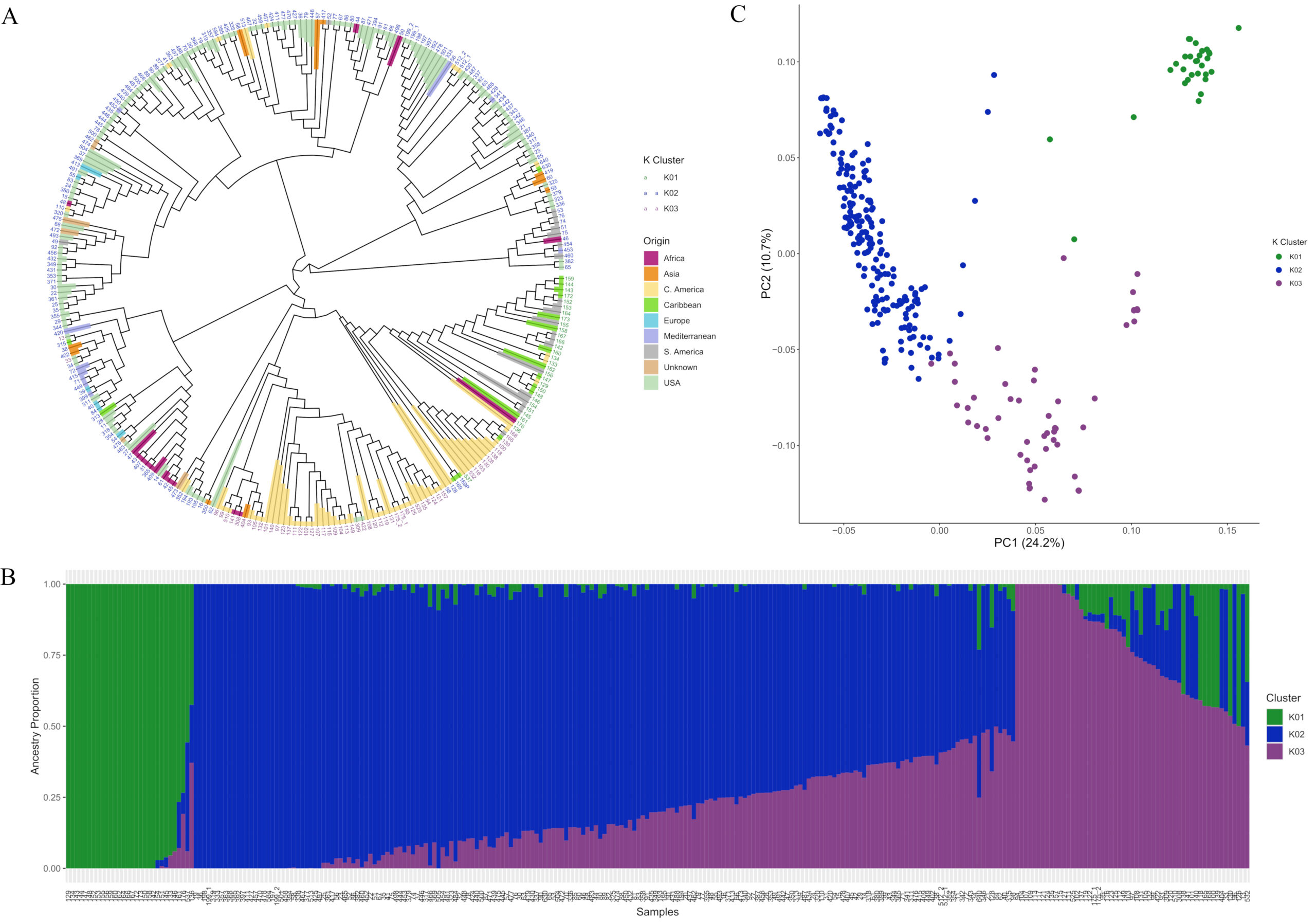
Leaf samples were collected from all the accessions and sent to a company, SGS Technologies, to do the genotype sequencing by running an 18K SNParray, we received SNPs for around 17K markers with around 10K that were homologous with unique markers and the other markers were either heterozygous and/or the marker matched multiple locations in the genome—using R version 4.4.2 and RStudio package called GAPIT (Wang and Zhang, 2021).
The field experiment included a field of 550 Upland Cotton varieties, for which leaf samples were collected to genotype the germplasm for eventual association mapping. The genotyping is planned to take place later in 2024. There were 24 genotypes selected for the above ground bag experiment. Only 2 replications could be completed with the lateness of the season, getting them planted, and the time and labor it took to fill the bags. The images of the roots were uploaded into the WinRHIZO software for the analysis of the traits with the ruler sticks set for image calibration. The results from the end of October, with the extraction of the bags, show some differences in the genotypes and amount of root length (Fig. 9). However, none can be statistically significant with the limited replications and missing data, making it impossible to validate these results statistically. A repeated experiment will need to be done. Only two replications could be done with the time it took to fill and the summer heat. The two replications also vary greatly because of the time difference in extracting the roots. Also, 12 samples were damaged in the hailstorm, which could account for some variation. The plants were also past their boll development in many of the plants because of the time it took to extract them. This could also have had an impact on the roots that were present. A lot was learned from this experiment; hopefully, the repeated experiment will not repeat the same mistakes.
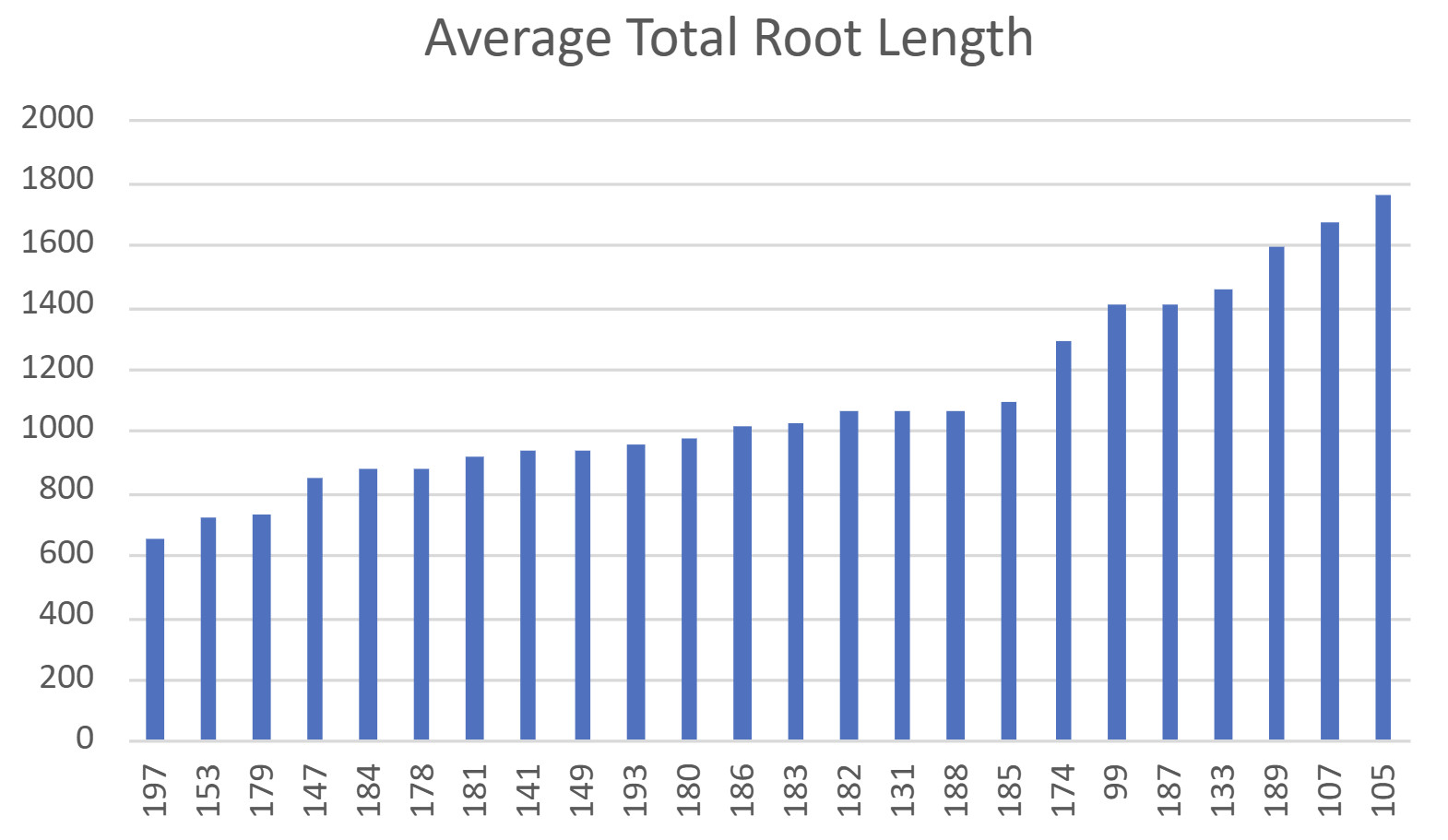
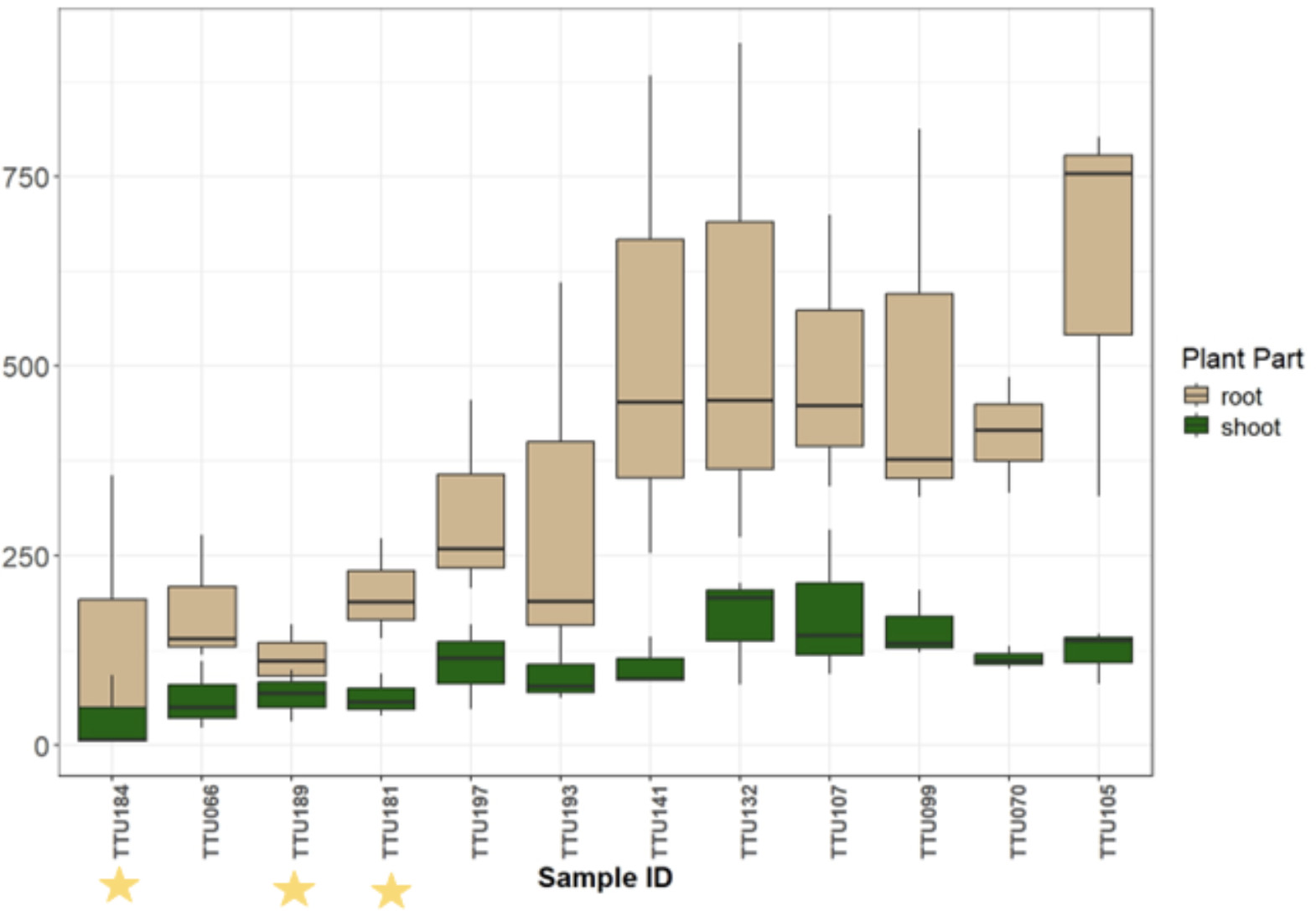
The next experiment was done in the greenhouse over the winter break and beginning of January 2024. The same bags were used with fewer accessions, but with three replications instead of two. The results in Figure 10 contain the total root length and the surface area of the shoots for each of the 12 accessions. The three elite cultivars have yellow stars to distinguish them from the other varieties. TTU193 is the Texas Marker 1 (TM1), and TTU197 is the Coker 312. These findings follow a similar pattern to the previous year’s greenhouse experiment in that the elite cultivars seemed to do poorer than TM1 and Coker312, and the landraces (132,099, 105, and 107) are continuing to outperform in root length compared to the elite cultivars and TM1 and Coker312. A couple of surprises for this are that TTU141 and TTU070 were expected to be lower compared to other accessions when looking at the previous year’s greenhouse data. However, it was observed that these accessions withstood some of the greenhouse’s insect pressures better than the elite cultivars did and could have had an advantage in their growth. Some of these landraces' more prolific root growth could potentially contribute to drought tolerance when placed in field conditions. To test this hypothesis, these accessions went on to objective two where field testing with drought stress was added.
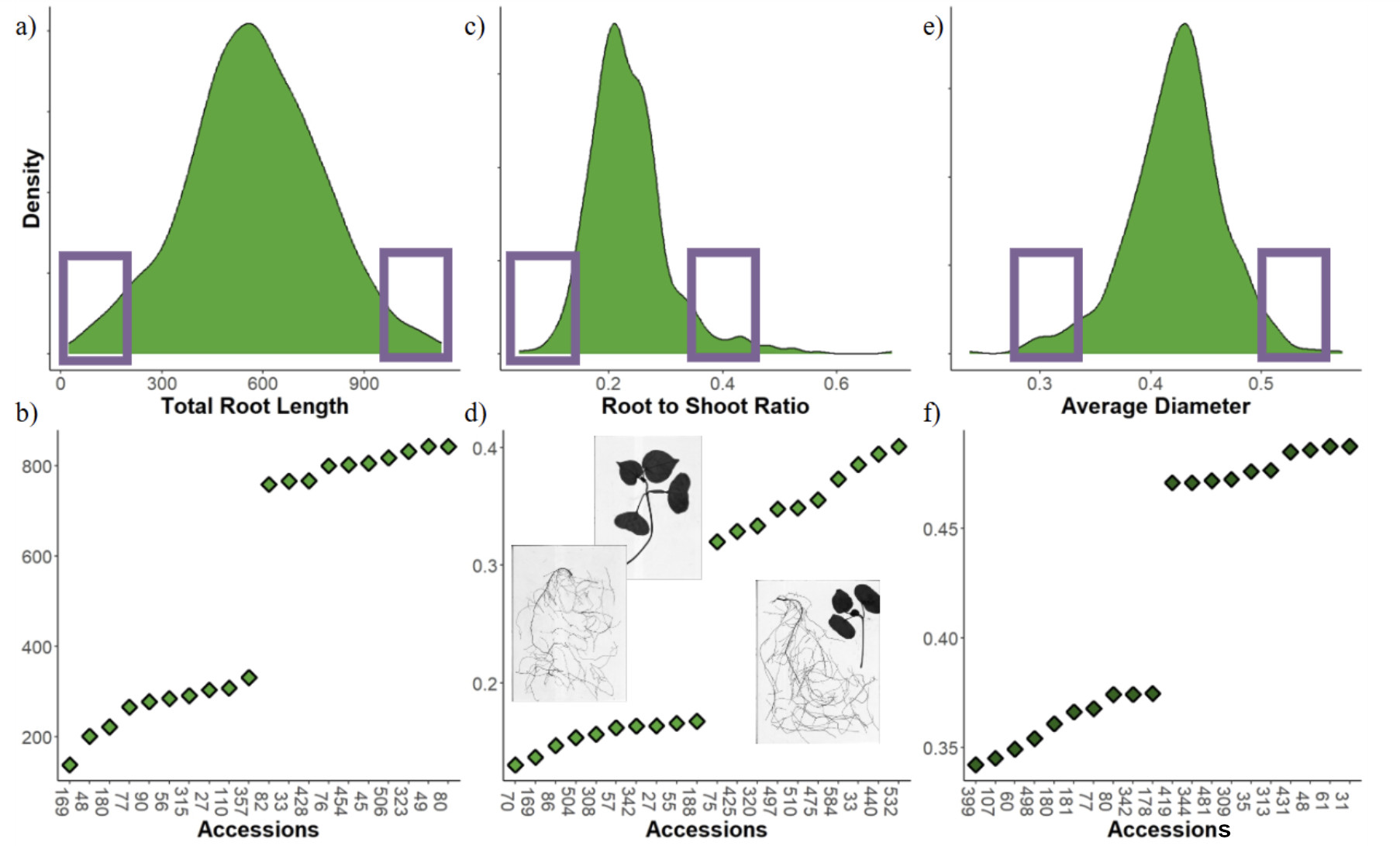
All accessions' total root length (TRL) shows a normally distributed pattern with a mean of around 570 cm of root length for 17 DAP. The range varies from 150 to around 900 cm, suggesting a large variation in the germplasm for total root length (Fig. 9). The colors represent the country of origin from which the accession seed was collected. Each color seems randomly spread out between high, middle, and low; suggesting that not just one origin has the highest root length.

Looking at the correlation between the RSA traits, most of them are positively correlated. The exceptions include the root to shoot ratio with several traits that can be negatively correlated and average diameter with the root length and average diameter with the root to shoot ratio with no correlation (Fig. 12). This is most likely because of the age of the seedlings being only two weeks old, most of the diameter is the same regardless of length or how tall the plant is. A good overall picture of each accession’s RSA can be determined by comparing these three traits.
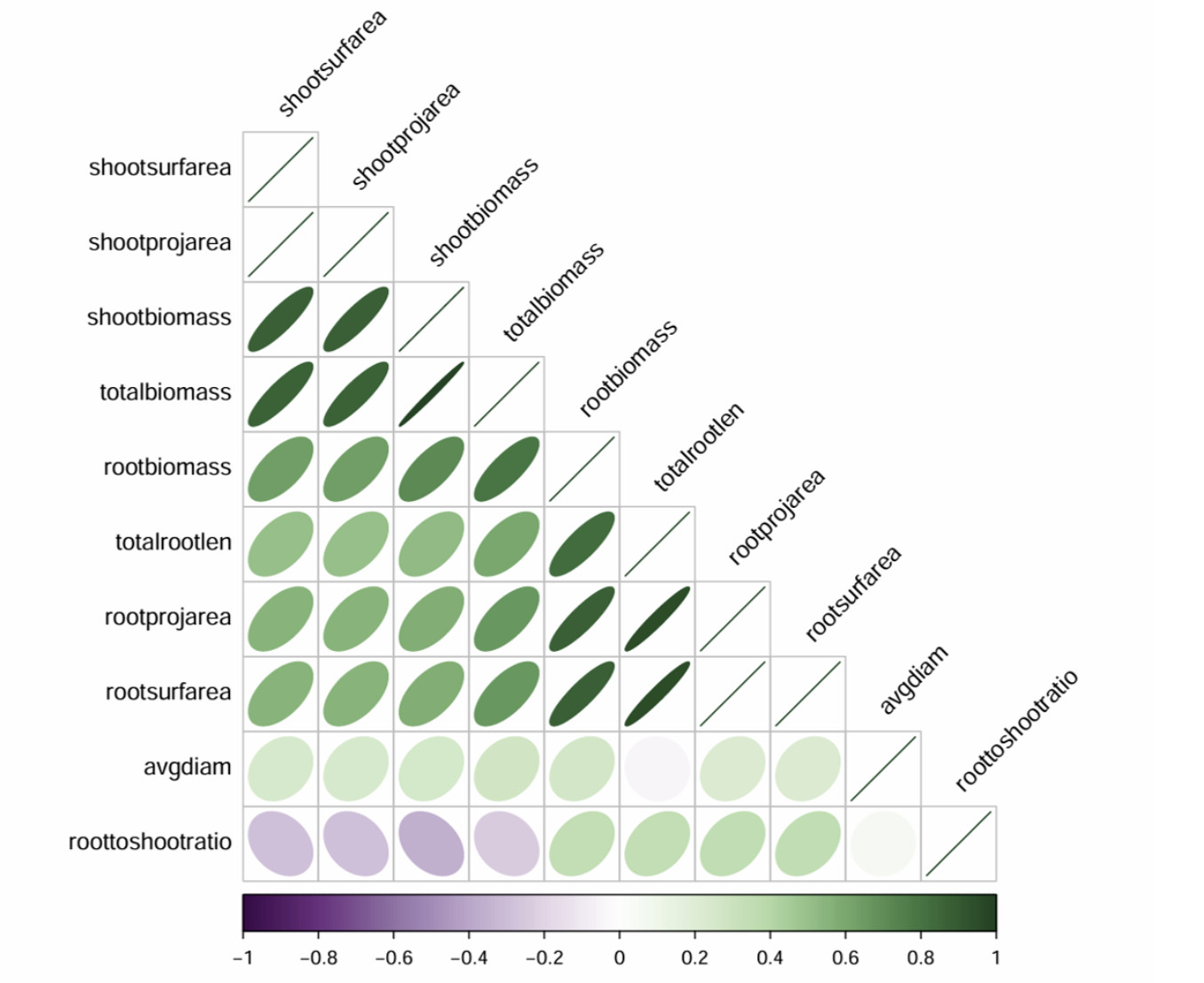
shootsurfarea = Shoot Surface Area (cm2)
shootprojarea = Shoot Projected Area (cm2)
shootbiomass = Shoot Dry Biomass (g)
totalbiomass = Total Dry Biomass (g) (root + shoot)
rootbiomass = Root Dry Biomass (g)
totalrootlen = Total Root Length (cm)
rootprojarea = Root Projected Area (cm2)
rootsurfarea = Root Surface Area (cm2)
avgdiam = Average Diameter (cm)
roottoshootratio = Root to Shoot Ratio (root biomass / shoot biomass)
When comparing the accessions with the most and least total root length, they may or may not also be found at the most and least root to shoot ratio or average diameter (Fig. 13). Accessions may have different strategies for drought stress response, by only looking at accessions really good at prolific root length other traits may be missed. Selecting some of the best and worst from each trait may be good to explore in additional experimentation with drought stress. Some elite varieties are found in the bottom ten of these traits. In the case of the average diameter the smallest ten is accession 80 which is also the number one spot for root length. Will 80 do well in drought conditions? To find out the same 212 accessions will be evaluated in objective 2.
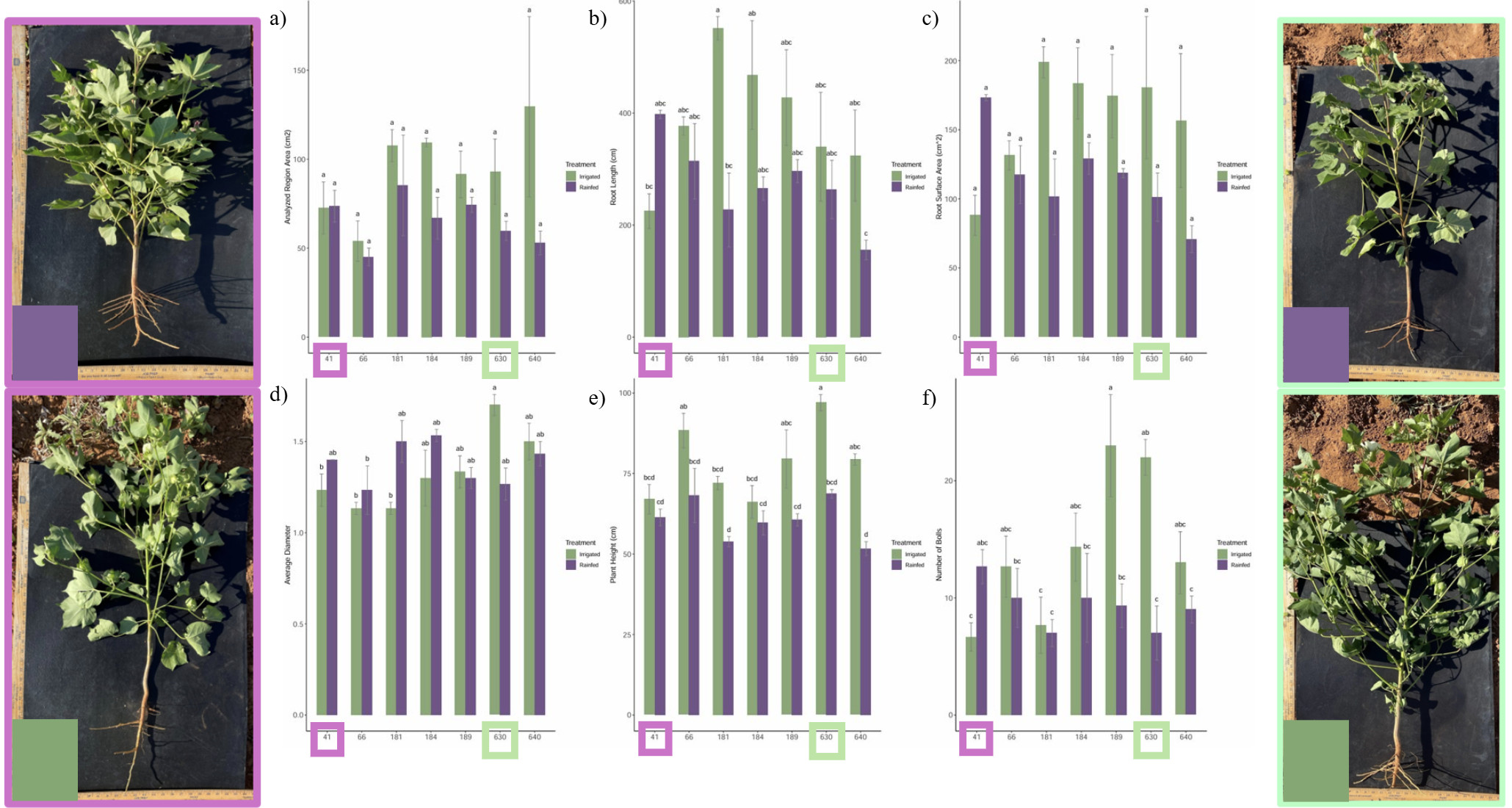
Some of the 12 accessions with more than 10 plants were selected to be pulled for just the crown of root systems to see if there was much difference in their root systems from rainfed to irrigated, and also to compare with each other. One sample that showed potential drought tolerance had an improved root system in the rainfed condition compared to the irrigated condition, while two accessions that showed drought tolerance had a reduced root length but a similar or increased root diameter. This could suggest that their drought tolerance came from above canopy physiological changes and/or the change in the diameter of the root could have helped with the drought stress response (Fig. 14). These three seemingly different strategies may support the finding that there are multiple strategies of drought stress tolerance and varying ways of changing the physiology and anatomy in response to that stress. Could there be a way to combine some of those varying strategies to improve or optimize the drought tolerance?
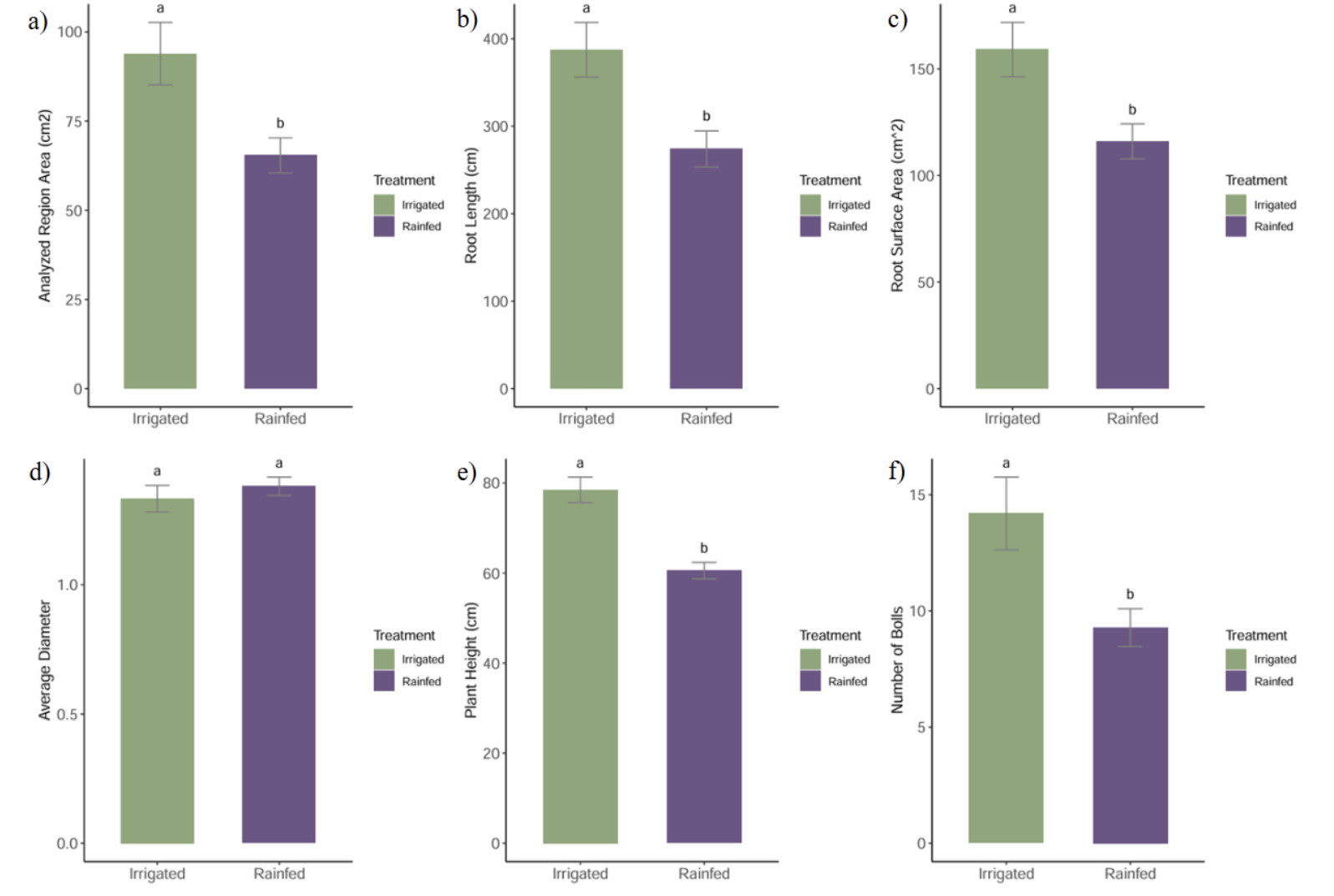
An ANOVA was performed to see if the treatments were significant for root traits, plant height, and number of bolls (Fig 15).
To get an overall look at the differences in growth and yield, a principal component analysis was conducted to see if any clusters or samples showed interesting patterns. Starting from the middle of the plot, as one goes to the left, the plant height decreases. As one goes to the right of the plot, the number of bolls, plant height, and weight of the plant increase. Closer to the bottom of the PCA grid is an increased boll weight (Fig. 16). There is a clear distinction between the irrigated (green) samples and the rainfed (purple) samples. Nearly cut vertically through the center. However, some purple samples perform just as well as the higher functioning irrigated samples. And some green samples perform low, close to the middle of the drought-stressed plants. If plants undergoing intensive drought stress can still have the same growth and yield as other irrigated plants, that could suggest having some drought stress tolerance.
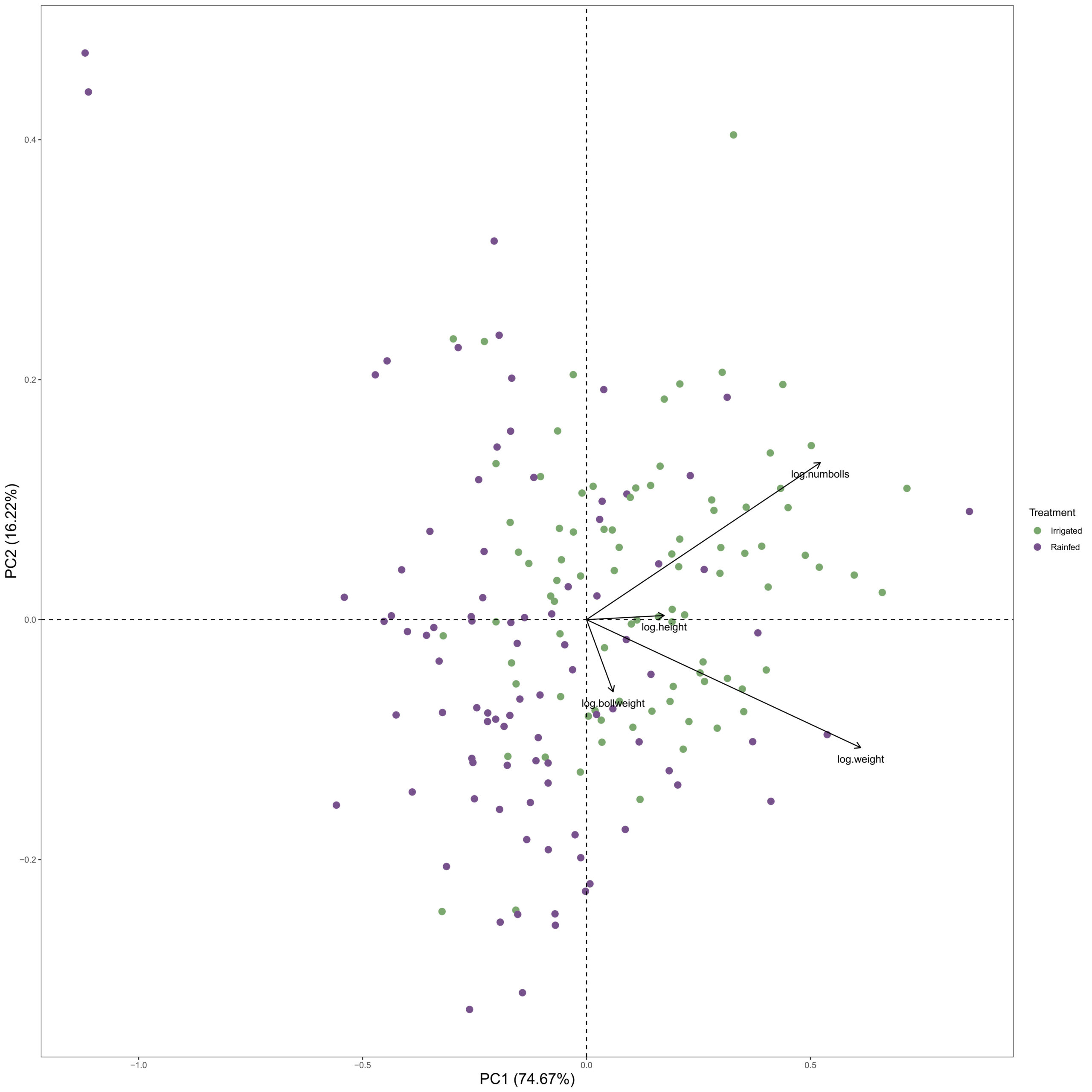
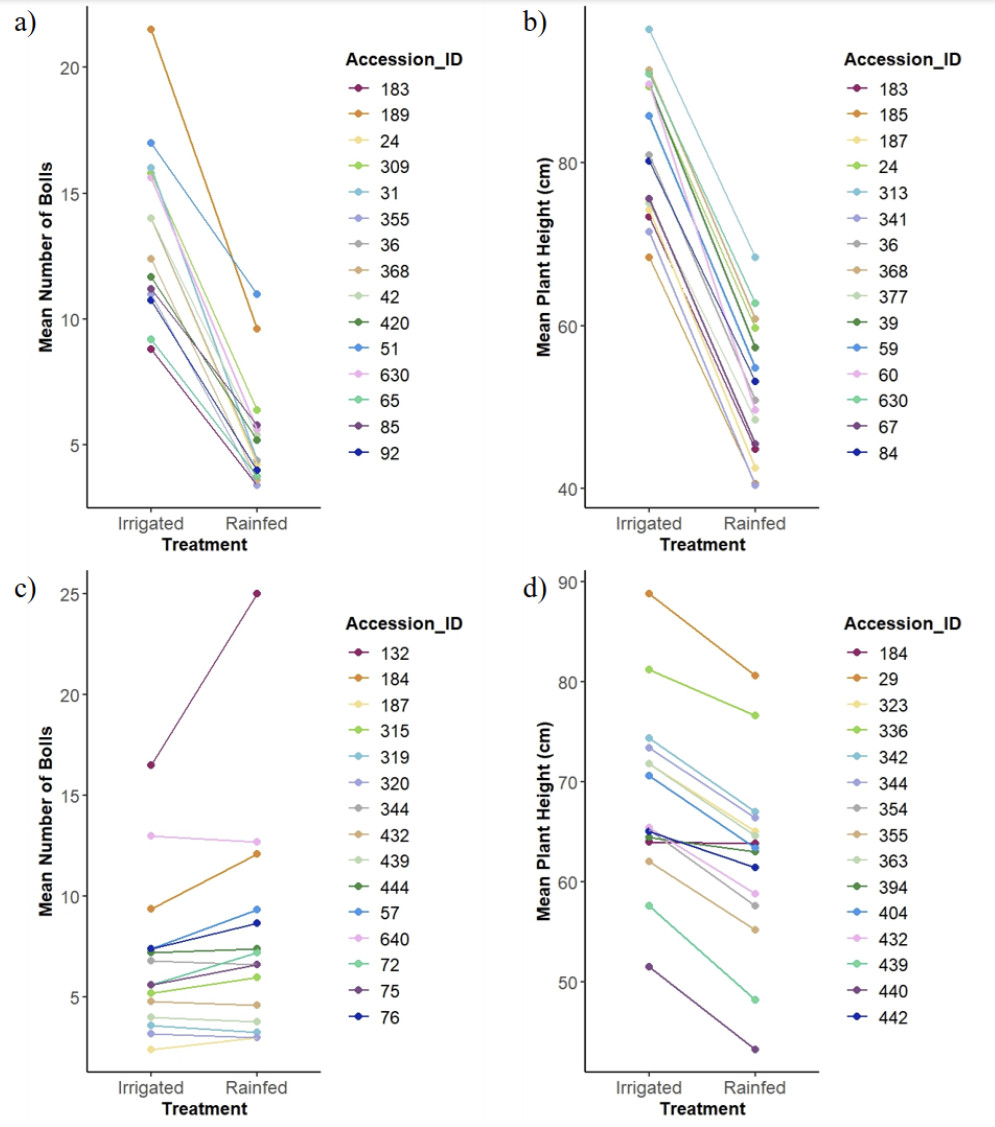
To get an idea of some of the accessions that performed similarly in both the irrigated and drought conditions, the means of the boll number and plant height were compared, and those with the greatest slope showed the greatest difference. Negative slopes suggest that drought hindered yield, while positive slope suggests that the accessions performed better in the drought condition compared to the irrigated condition. A small or near horizontal line suggests that they performed nearly the same in both conditions (Fig. 17).
These results together can work to select high and low performance with RSA traits and field performance under drought stress conditions. This selection of contrasting lines can then continue to the next level of experimentation with increased replications, added drought stress in the greenhouse, and another year of field experimentation. In addition to all the previous parameters, physiological data will be critical to measure in the upcoming experimentation. Physiological measurements like stomatal conductance, chlorophyll levels, and relative water content of the leaves give a greater understanding of the condition of the plant and how the physiology plays a role in drought tolerance, along with what the accession maintains.
Objective 3: Analysis of phenotypes and genotypes through genome-wide association mapping
By collecting leaf samples of all the accessions and sending them to a company, SGS Technologies, to do the genotype sequencing by running an 18K SNParray, we received SNPs for around 17K markers with around 10K that were homologous with unique markers and the other markers were either heterozygous and/or the marker matched multiple locations in the genome. Using R version 4.4.2 and RStudio package called GAPIT (Wang and Zhang, 2021). Several traits had markers that showed significant QTL peaks (Fig. 19).
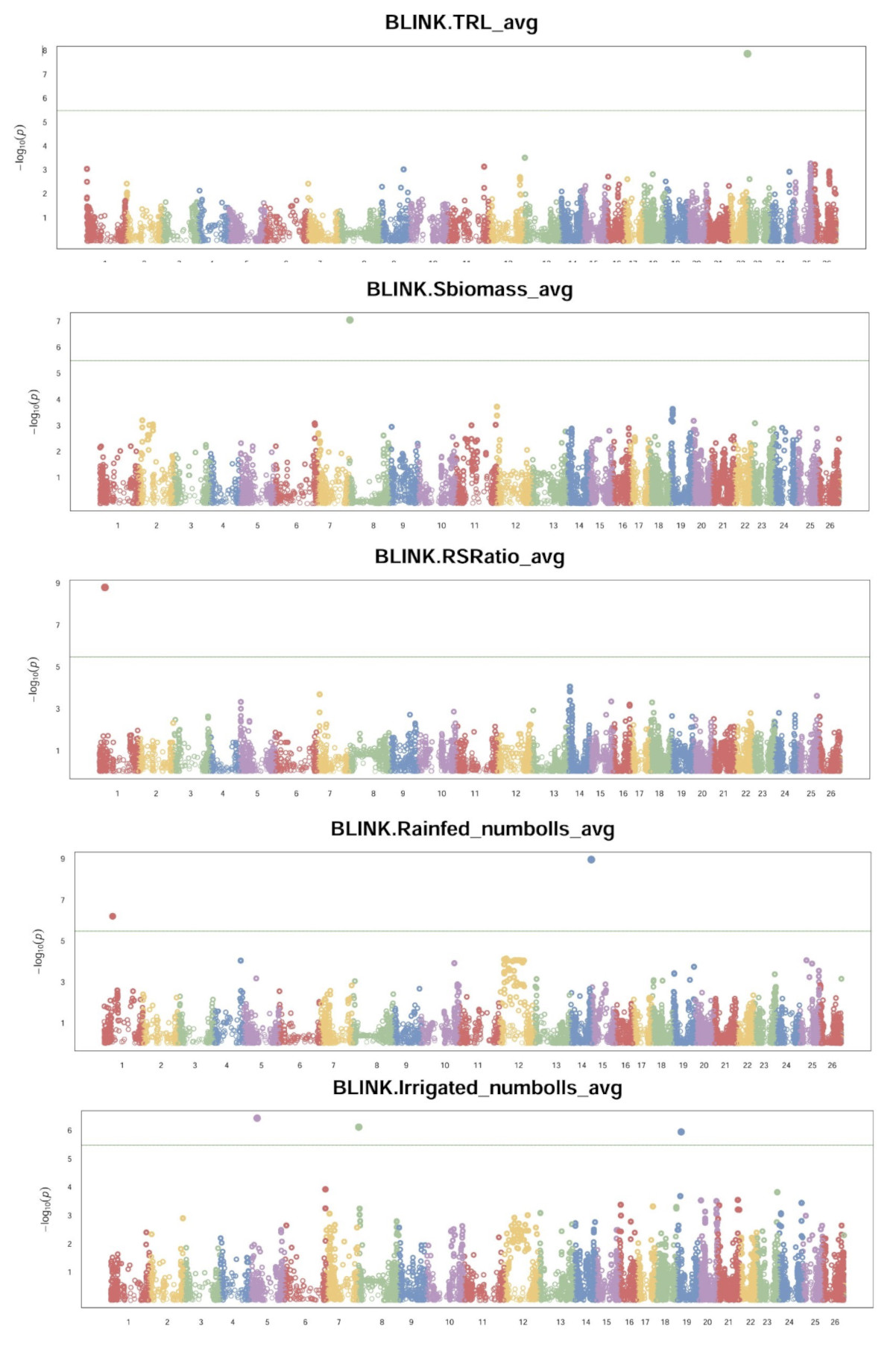
Because this data is very recent, more time is needed to analyze and process it all before conclusions can be drawn about these potential QTLs.
Educational & Outreach Activities
Participation Summary:
Presented my research at the 10th Annual Water College in January 2024. Farmers, sponsors, and educators were present at the event of over 200 hundred people.
Lamb, M. L. (2024, January 24). Optimizing cotton's root system to improve response to water-deficit stress [PowerPoint slides]. Institute of Genomics for Crop Abiotic Stress Tolerance, Texas Tech University. https://texastechuniversity-my.sharepoint.com/:p:/g/personal/miclamb_ttu_edu/Ede1MWpL2SxNu8MjQb-bWEYBcdg5L-_jzAUcg3wI-n_O8Q?e=dWC8qk&nav=eyJzSWQiOjI1NiwiY0lkIjoxMDk4NTcyMjJ9
Presented at the Agricultural Water Sustainability Summit, Lubbock, Texas, on August 9, 2024, a few hundred farmers, ranchers, and agricultural professionals attended the conference. My presentation was on the last day, towards the end of the conference, so only 12 people were present for my presentation.
I presented at the Beltwide National Cotton Conference on January 16, 2025, at the Center New Orleans, Louisianna. Although thousands of farmers/ranchers and agricultural professionals participated, around 50 were in attendance for my presentation.
Presented my field research at the Quaker Farm Field Day August 2024 with around 25 farmers, or Agricultural professionals
Project Outcomes
Suppose we can figure out more about the roots, how they are related to shoot traits, and the genetic relationships with the roots. In that case, It will save future generations time, labor, and money to develop their crops with a better understanding of the Root System Architecture. Because it is tough to view the roots to know what traits are being bred for and what root traits will be beneficial or not in different abiotic stresses, this project's goal is to get additional understanding of methods of how to assess the RSA and also do it in an economically inexpensive way to better our understanding of the genetic mechanisms of the RSA.
The data suggest that the cotton germplasm has a significant variation in RSA traits, and various accessions are more tolerant of drought than others. There could be strategies that include altering the root system during drought stress to help the plant endure the periods of drought without suffering as much yield loss. By comparing the results of this study, we can select accessions that may have desirable RSA traits that can be further studied and understood to be applied to future studies and breeding programs. Further analysis of the GWAS results to further the understanding of potential genes that play a role in those traits will also help determine how to optimize the traits for drought tolerance. Accession TTU 132, TTU 184, TTU 80, and a few others that show promise of drought tolerance and those that show drought sensitivity can be narrowed down into more focused experimentation that can lead to even greater understanding of RSA's role in drought tolerance for cotton.
We are still learning as we continue these few months since September 2023. I have been learning how much work is required to learn something unknown and also how much knowledge that comes from so many other people before is needed to build upon each other. I am excited for the next year of learning and hope to come up with new information to better serve society and agricultural partners.
It is recommended that additional greenhouse testing with selected accessions do additional drought testing and repeated year experiments to further validate results. It is also recommended that additional investigation into the GWAS results and further study of any QTLs and candidate genes to implement into experiments for the following year.





 B)
B) 