Progress report for GS24-303
Project Information
Powdery mildew is a major fungal disease that causes significant yield and quality losses in squash (Cucurbita spp.) production. Conventional growers rely on routine fungicide application to manage the disease, costing an average of $350/ acre per season. Organic growers have limited control options leading to heavy yield losses when the disease pressure is high. The availability of squash cultivars resistant to powdery mildew would reduce yield losses for growers and promote environmental stewardship through reduced pesticide application. However, resistance to powdery mildew in most commercial squash cultivars is lacking or, at best, weak, necessitating complementary and substantial fungicide applications to ensure profitability. Recently, we conducted a single greenhouse screen of the USDA core squash collection (n= 207) in Florida and identified several novel sources of genetic resistance to powdery mildew. Using the screening data, we performed a preliminary genome-wide association study (GWAS) analysis, revealing potential markers that could be useful for marker-assisted selection. Building upon this preliminary work, we propose to i) conduct a second greenhouse screening of the entire core collection and validate the new resistance sources, ii) perform a comprehensive GWAS analysis using multi-screening phenotype data and validate molecular markers linked to resistance, iii) transfer powdery mildew resistance into elite squash cultivars using marker-assisted selection, and iv) characterize the inheritance mechanism of resistance in novel resistant sources. The novel powdery-mildew-resistant germplasm will be evaluated for yield and horticultural performance under field conditions, and the findings will be communicated to growers through extension outreach.
- Conduct a second greenhouse screening of the USDA squash core collection and validate new resistance sources: It builds on preliminary findings to confirm resistance traits under controlled conditions, ensuring that identified resistance is robust and reproducible.
- Conduct a comprehensive GWAS analysis using multi-screening phenotype data and validate molecular markers linked to resistance: The rationale here is to utilize advanced genomic tools to decipher the genetic basis of resistance to powdery mildew in squash. Unlike traditional breeding, which relies on multiple growing seasons to confirm traits, thus utilizing many resources (soil, fertilizers, manual labor), GWAS enables quicker and more accurate identification of resistant genotypes.
- Transfer powdery mildew resistance into elite squash cultivars using marker-assisted selection: This objective seeks to apply the findings from genomic analyses to practical breeding. The justification lies in its direct impact on developing commercially viable, disease-resistant squash varieties, reducing reliance on fungicides, and lowering production costs.
- Characterize the mode of inheritance in the novel resistant sources: Understanding how resistance traits are inherited is fundamental for effective breeding. This objective aims to inform breeding strategies by elucidating the genetic mechanisms through which resistance is passed from one generation to the next, which is critical for predicting the behavior of these traits in breeding programs and ensuring the stability of resistance in future cultivars.
Research
Objective 1: Conduct a second greenhouse screening of the USDA squash core collection and validate new resistance sources.
Under this objective, greenhouse screening of the USDA C. pepo core collection (n= 207) accessions for powdery mildew resistance will be conducted. The experiment will be carried out at the University of Florida- Tropical Research and Education Center and will follow a randomized complete block design consisting of three replications and five plants per accession (n= 15). The inoculation of plants will follow a standard protocol for powdery mildew screening in the greenhouse (Zhang et al., 2021). Briefly, powdery mildew inoculum will be obtained from the leaves of susceptible and heavily infected squash plants. The leaves will be agitated in a solution comprising distilled water and the surfactant Silwet L-77 at a concentration of 250 µL/L. Subsequently, a spore suspension containing powdery mildew conidia at a concentration ranging from 104 to 105 spores/ ml will be applied uniformly across the plant's surface using a spray bottle. Disease severity will be assessed at regular intervals using a 0-100% rating scale, where 0 corresponds to clean/resistant plants, and 100% corresponds to highly susceptible plants. The disease rating will be taken for the following parameters: top 4th leaf surface, bottom 4th leaf surface, stem above 4th leaf, stem below 4th leaf surface, and whole plant rating. The comprehensive phenotypic data collected from these greenhouse evaluations will be used to confirm the resistance in novel accessions and to conduct a comprehensive GWAS analysis.
Objective 2: Conduct a comprehensive GWAS analysis using multi-screening phenotype data and validate molecular markers linked to resistance
The genotype (re-sequencing) data for C. pepo core collection is publicly available through the Boyce Thompson Institute, Cornell University (Fei Lab). The genotype data consisting of more than 4 million single nucleotide polymorphisms (SNPs) will be converted to a numeric text (.txt) format, followed by the execution of the Genome Association and Prediction Integrated Tool (GAPIT) in R software through the University of Florida HiPerGator computing infrastructure. Three statistical models (MLM, FarmCPU, and Blink) will be deployed to detect genomic loci significantly associated with powdery mildew resistance. A threshold of p<0.001 will be used to determine significant associations between SNPs and powdery mildew resistance. Genomic regions harboring significant SNPs will be blasted against the C. pepo reference genome to identify the putative physical positions and candidate genes. SNP markers will be converted into KASP (Kompetitive Allele Specific PCR) assays and validated in independent segregating populations before use in marker-assisted selection.
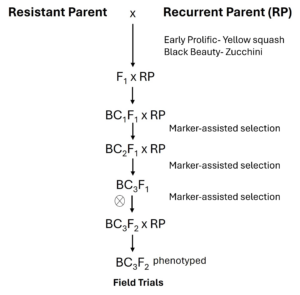
Objective 3: Transfer powdery mildew resistance into elite squash cultivars using marker-assisted selection. Under this objective, a marker-assisted backcross breeding scheme using SNP (KASP assays) markers linked to powdery mildew resistance in the novel accessions will be implemented to transfer novel alleles into elite C. pepo genetic background (Figure 2). Briefly, the resistant parents will be crossed with elite but susceptible C. pepo cultivars—Early Prolific Straightneck (EP) and Black Beauty (BB). F1 hybrids will be germinated and genotyped using molecular markers linked to resistance to confirm the presence of resistance genes. F1 individuals exhibiting resistance and desirable traits will be backcrossed with elite cultivars (EP & BB). Several backcrosses will be performed, ensuring progressive introgression of resistance and recovery of elite fruit traits. Greenhouse/ field trials will follow to evaluate the horticultural performance and resistance of the backcross progeny.
Objective 4: Characterize the mode of inheritance in the novel resistant sources.
Under this objective, the inheritance mechanism of powdery mildew resistance in the novel resistant accessions will be determined. A controlled cross between the resistant and susceptible (Early Prolific Straightneck and Black Beauty Zucchini) parents will be made to generate F1 seed. The F1 hybrids will be self-pollinated to produce F2 populations and backcrossed with the respective recurrent parents (RP) to develop corresponding BC1F1 populations. The parents, F1, F2, and BC1F1 populations will be phenotyped following the previously described protocol (see objective 1). A χ2 test (McHugh, 2013) will be used to compare segregation ratios for each population with hypothetical segregation patterns to determine possible inheritance mechanisms for powdery mildew resistance in the novel resistant accessions.
The project is underway according to the proposed timeline.
Objective 1: Conduct a second greenhouse screening of the USDA squash core collection and validate new resistance sources
- The germplasm collection of C. pepo accessions (n=207) is currently in the greenhouse for a second phenotype screening.
Objective 2: Conduct a comprehensive GWAS analysis using multi-screening phenotype data and validate molecular markers linked to resistance
- Based on the first greenhouse screening, the GWAS analysis has been conducted using GAPIT and different statistical models such as BLINK, FarmCPU, and MLM which revealed potential markers on several chromosomes such as for top 4th leaf (Chr 6, 11, 13 and 19), bottom 4th leaf (Chr 1, 9, and 12) stem above 4th leaf (Chr 1, 4, 14, and 16), stem below 4th leaf (Chr 4, 11 and 12) and whole plant (Chr 1, 3, 9, and 15), with co-located loci on Chr 15 suggesting linkage/pleiotropy. Currently, candidate gene search is underway, and the target regions will be validated for the population of interest through KASP markers.
Educational & Outreach Activities
Participation summary:
Consultations:
- Consultation on Disease Inoculation and Evaluation Methods for Starke Ayres (2024): Provided guidance on plant disease inoculation and evaluation techniques to Wilmarie Kriel, Plant Pathology Manager at Starke Ayres, including comprehensive recommendations for implementation in controlled environments.
Journal articles:
The following research review encompasses the preliminary literature review on the subject. Submitted before receiving the grant but relevant to the project:
- Sabharwal, P., Thakur, S., Shrestha, S., Fu, Y., & Meru, G. (2024). Breeding and genetics of resistance to major diseases in Cucurbita—A review. Crop Science. https://doi.org/10.1002/csc2.21358
Presentations:
- Oral Presentation at XIII Eucarpia Cucurbit Breeding and Genetics 2024, Naples, Italy- "Exploration of Novel Genetic Resistance to Powdery Mildew in Cucurbita pepo Using Genome-Wide Association Studies"
- Oral Presentation at ASHS 2024, Honolulu, Hawaii- "GWAS analysis of USDA Cucurbita pepo germplasm for powdery mildew resistance"