Progress report for GS24-312
Project Information
Tomato spotted wilt virus (TSWV) is one of the chronic pests listed in the priority list identified by stakeholders and published by the Southern Region Information Exchange Group for IPM (SERA3) members and Integrated Pest Management coordinators from the southern region of the United States. Tomato spotted wilt disease (TSWD) is broadly managed by integrating disease management tools using host-mediated resistance, vector management, and cultural practices. Even with devised management strategies, the disease incidence remains high in peanuts and tobacco. In contrast, the emergence of resistance-breaking isolates in tomato and pepper has resulted in the decline of resistant varieties’ effectiveness as a management strategy. With this study, we aim to develop a Spray-Induced Gene Silencing (SIGS) technique against virulent strains of TSWV. This technique includes the production of double-stranded RNA (dsRNA) molecules, capable of inducing a natural plant defense mechanism against viruses. The dsRNA will be designed against the NSm (encode movement protein) and Gn/Gc (encode glycoproteins) genes of TSWV. The findings from this study may yield a critical component for formulating various products applicable to different crop hosts. Given the widespread occurrence of RNAi, introducing dsRNA targeting TSWV can potentially confer immunity across all its host species, assuming successful uptake of the dsRNA by the host organisms. Due to its precise nature, it promotes green practices by avoiding off-target effects. As we are on the verge of losing important management strategies for TSWD, SIGS developed against TSWV will provide an effective approach to sustainable agriculture.
The aims of this project are:
- Evaluation of symptom severity and TSWV titer on application of in vitro produced dsRNA against NSm and Gn/Gc genes of TSWV, in tobacco plants.
- In vitro production of dsRNA against NSm and Gn/Gc genes of TSWV.
- TSWV inoculation and application of dsRNA on the tabacum plants.
- Evaluation of symptom severity and TSWV titer in inoculated and systemic leaves.
- Evaluating dsRNA movement and small RNA (sRNA) population using High Throughput Sequencing (HTS) in the tabaccum on different days post-application.
- Disseminating the research to the stakeholders for disease management.
Expected Outcome.
Objective 1:
Two different dsRNA molecules will be generated in the first objective. These dsRNA will have sequence similarity to the conserved region of the NSm and Gn/Gc, respectively. The second objective will help to decipher the efficacy of these dsRNA molecules against TSWV. Low disease incidence and severity are expected to be observed upon the treatment of inoculated plants with dsRNA. Further, viral titer in both inoculated and systemic leaves is also anticipated to decrease. This will help us to conclude the performance of dsRNA against NSm and Gn/Gc genes of TSWV at different DPIs.
Objective 2:
The administered dsRNA should move to other parts of the plants for better functioning. So, this project also includes the objective to evaluate its movement. The dsRNA applied is expected to be detected on the systemic leaves above the inoculated leaves. Further, high diversity in the siRNA population is expected. This will keep the concentration of any one siRNA low and help to reduce any off-target effects.
Objective 3:
Stakeholders will receive research results through the UGA extension network and presentations at county agents and growers’ meetings led by the UGA Cotton and Peanut team. Additionally, the results will be shared with growers, consultants, and other stakeholders during the annual GA-FL tobacco tour organized by the Georgia Commodity Commission on Tobacco and the peanut tours conducted by the GA Peanut Commission in the summer and fall. Overall, this objective will provide the crucial information to carry forward this research according to the stakeholders’ needs.
Research
In our 2021 study, we applied dsRNA against Tomato Spotted Wilt Virus (TSWV) on tobacco plants under controlled conditions. We induced resistance in tobacco plants by targeting the N and NSs genes. We observed the movement of dsRNA from inoculated leaves to systemic leaves using semi-quantitative RT-PCR. Both dsRNAs reduced viral titer and symptom severity. Notably, dsRNA against the N gene resulted in only 16%, and dsRNA against NSs led to 54% of disease incidence. Overall, both dsRNAs outperformed no treatment controls, with dsRNA against N showing superior results (fig 4) (Konakalla et al, 2021).

We explored two approaches on tobacco plants: curative (TSWV inoculation followed by dsRNA application) and preventive (dsRNA followed by TSWV inoculation), targeting the N and NSs genes of TSWV. Statistical analysis reveals a substantial reduction in virus titer after treating inoculated plants with dsRNA. In both approaches, dsRNA against the N gene outperformed the NSs gene. Notably, preventive dsRNA application against the N gene significantly decreased TSWV titer in both inoculated and systemic leaves (figs 5 and 6) (Unpublished).
To further expand this research, we are aiming to target the NSm and Gn/Gc of TSWV. This will help to better understand the essentiality of these genes in the pathogenicity of TSWV. All the experiments will be conducted in the Crop Virology laboratory on the Tifton campus of the University of Georgia
The method for this project would be the following:
Objective 1:
- To find the conserved regions to construct dsRNA against NSm and Gn/Gc genes of TSWV.
Sequences of NSm and Gn/Gc will be retrieved from NCBI GenBank as well as from different isolates collected by our lab members from various counties of Georgia, US. MEGA-X software will be used to align multiple sequences. Conserved regions of each gene will be used to design primers (Konakalla et al, 2021).
- To produce dsRNA molecules targeting the NSm and Gn/Gc genes of TSWV.
dsRNA molecules will be produced through a two-step PCR approach followed by in vitro transcription. To begin with, total RNA from TSWV-infected N. tabaccum plants will be extracted using the Quick RNA miniprep kit (ZYMO Research, US) according to the manufacturer’s instruction. 2 μg of total RNA will be used as a template for cDNA synthesis using SuperScript™ IV Reverse Transcriptase (Thermo Scientific, US). Viral genes will be amplified with specifically designed primers. The T7 RNA polymerase promoter sequence will be added to the 5’ end of both forward and reverse primers. PCR will be performed using about 100 ng of cDNA as a template. To synthesized sense and antisense viral derived ssRNA, in vitro transcription system named T7 RibomaxTM Express large-scale RNA production kit (Promega, Madison, US) will be used with 1 microgram of purified DNA template. After removing all the DNA from the reaction mix, dsRNA will be obtained by mixing the specific sense and antisense ssRNA. The formation of dsRNA will be confirmed by Mung Bean Nuclease treatment (NEB, US). As a negative control, dsRNA against the coat protein of cucurbit chlorotic yellows virus (CCYV) will be used.
- To apply dsRNA molecules along with TSWV onto the tabacum.
TSWV inoculum stock will be prepared from the N. tabacum plants inoculated with isolates obtained from the field samples. Each inoculation will be prepared by homogenizing 1 g of infected plant tissue in 25 ml of phosphate inoculation buffer (pH 7.0). The plants will be inoculated at the four-leaf stage. Each leaf will receive 50 μl of inoculation buffer with TSWV on the adaxial side. 10 μg of dsRNA will be added during inoculation in treatment. There will be a total of three replicates with 10 plants for treatment (TSWV with dsRNA), 5 plants as untreated control (only TSWV), 5 as negative control (TSWV with dsRNA against coat protein of CCYV), 5 as a mock plant (only inoculation buffer with dsRNA) and 5 plants as negative control (only with inoculation buffer). This experiment will be repeated three times using both dsRNA against NSm and Gn/Gc genes of TSWV.
- To measure the symptom severity and viral titer in both treated and untreated plants.
Three samples from each replicate will be collected at 6, 9, and 12 DPI. TSWV titer will be measured using qPCR using primers for the N gene of TSWV. The plants will be observed till 30 DPI. A severity scale of 0 to 5 will be used where, 0= no symptom, 1= chlorotic and necrotic spots in inoculated leaves, 2= initiation of necrotic spots in systemic leaves, 3= spread of necrotic spots in systemic leaves, 4= necrotic spots and vein necrosis in systemic leaves, and 5= stunted growth with severe necrosis in systemic leaves.
Objective 2:
- To evaluate dsRNA movement inside tabacum plants.
Two leaf disks from each inoculation and the systemic leaf from the mock plants will be taken at 1-, 3-, 6-, and 9 days post-inoculation (DPI). Three plants will be used from each time point. The leaves will be washed with Triton X-100 (0.05%) and water to remove residual dsRNA from the leaf surface. cDNA will be performed using random primers and dsRNA will be quantified using q-PCR with the help of primers specific for dsRNA against NSm and Gn/Gc genes (Tabein et al, 2020).
- To evaluate small RNA (sRNA) populations originating from dsRNA against NSm and Gn/Gc genes.
The samples used for dsRNA movement at 1 DPI will be subjected to high throughput sequencing for sRNA analysis. Small RNA libraries preparation and sequencing will be performed by BGI genomics (China). The Qiagen CLC genomics workbench will be used to process the raw data.
Objective 3:
We’ll disseminate the results to stakeholders through diverse platforms like UGA extension networks, county agents, growers’ meetings, and field tours, along with prioritizing inclusion, and equity.
Project report till: March 31, 2025
Tomato spotted wilt virus (TSWV) causes tomato spotted wilt disease (TSWD), a major threat to horticultural and row crops in the southern United States. Therefore, it is listed as a chronic pest in the priority list identified by all the stakeholders. TSWD incidence is reduced by integrated disease management tools. Despite such strategies, the disease remained variable across decades in the Southeast. With this study, we aim to develop the Spray-Induced Gene Silencing (SIGS) technique against virulent strains of TSWV. This technique includes the production of double-stranded RNA (dsRNA) molecules targeting genes of TSWV, which will be used to generate plant immunity.
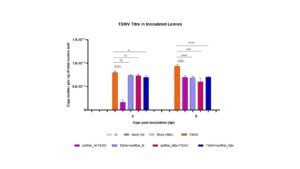
Till now, we produced dsRNA against NSm and Gn/Gc genes of TSWV. We have applied both the dsRNA with TSWV inoculum to the tobacco (Nicotiana tabacum) plants. The plants are kept under controlled conditions and will be monitored till 30 days post-application (DPA). Till 12 DPA, there is less disease incidence and severity in plants provided with dsRNA against the NSm as compared to Gn/Gc gene. Further, more replications are required to provide any concluding results.