Final report for GW22-238
Project Information
The USA produces ≤20% of the world’s spinach seed, with western Washington and Oregon the only region of the USA suitable climatically for spinach seed crops. Pathogen-free, high quality seed is needed to meet the demand for fresh and processing spinach crops, particularly baby leaf crops planted at 3-4 million seed/acre. Stemphylium leaf spot of spinach, caused by the fungi Stemphylium vesicarium and S. beticola, has become difficult to manage following widespread development of fungicide resistance in populations of S. vesicarium. This study will use population genetics to clarify the relative contribution of infected seed lots as inoculum for disease outbreaks. Populations of S. vesicarium from seed lots grown in major spinach seed production regions of the world, including the USA, will be compared to assess the probability that S. vesicarium isolates are moved among regions of seed production and sales through global seed trade. Populations of S. vesicarium from baby leafy crops with Stemphylium leaf spot will be compared with populations from seed lots planted in these fields. The results and knowledge generated recently on fungicide resistance, spinach cultivar resistance, and epidemiology of the disease will be updated online resources, including the PNW Plant Disease Management Handbook and the PNW Vegetable Extension Group. An extension bulletin will be published and results shared at the Western Washington Seed Workshop, Texas Spinach Field Day, and WSU Mount Vernon Field Day in 2023. Reports will be shared with seed growers, companies, and at the 2023 American Phytopathological Society Annual Meeting.
- Quantify the relative abundance of Stemphylium species pathogenic on spinach that occur on spinach seed lots.
- Asses the role of global spinach seed trade in moving isolates of vesicarium, causal agent of Stemphylium leaf spot of spinach, by characterizing populations of S. vesicarium genetically from seed lots grown in the five major spinach seed-producing countries of the world.
- Determine the relative significance of seedborne S. vesicarium as an inoculum source for outbreaks of Stemphylium leaf spot by comparing isolates of the fungus from leaves of baby leaf spinach crops symptomatic with the disease to isolates from seed lots used to plant these spinach crops.
- Disseminate results of this study and other recent knowledge generated on this disease at WSU to spinach seed producers, fresh market and processing producers, vegetable seed companies, other scientists that work with spinach, and the general public through oral, poster, and written presentations at fields days, a seed grower workshop, and an academic conference, and by updating the information on this disease available in the online Pacific Northwest Plant Disease Management Handbook (https://pnwhandbooks.org/plantdisease/host-disease/spinach-spinacia-oleracea-stemphylium-leaf-spot) and the spinach photo gallery of the Pacific Northwest Vegetable Extension Group
The first objective is to determine diversity of Stemphylium spp. associated with spinach seed, and prevalence of S. vesicarium. Forty single-spore isolates of S. vesicarium from will be generated from each of 10 spinach seed lots from Aug. to Oct. 2022. From Sep. to Nov. 2022, DNA will be extracted and the cmdA locus sequenced from isolates to confirm Stemphylium species.
The second objective is to determine potential impact of global spinach seed movement on genetic diversity of Stemphylium vesicarium, and the third objective is to determine significance of infected spinach seed as a primary inoculum source by comparing isolates of S. vesicarium from spinach seed vs. infected leaves from crops planted with infected seed lots. Tasks for these two objectives will be done in parallel. PCR products will be sequenced from a of subset of S. vesicarium isolates with 11 SSR markers to confirm they work for spinach-pathogenic isolates of the fungus from Sep. to Oct. 2022. Genotypes of S. vesicarium isolates will be characterized by sending SSR PCR products for capillary electrophoresis from Oct. to Dec. 2022. SSR capillary electrophoresis data will be processed from Nov. 2022 to Jan. 2023. Genotypes of S. vesicarium populations will be analyzed statistically from Jan. to Apr. 2023.
The fourth objective is the education and outreach activities to share knowledge with scientists, spinach producers, seed company personnel, and the general public. The PNW Plant Disease Management Handbook and PNW Vegetable Extension Group photo gallery on Stemphylium leaf spot of spinach will be revised from Oct. to Dec. 2022. In Jan. 2023, Kayla will present to seed producers and seed company representatives at the Western WA Seed Workshop. In Feb. 2023, Kayla and Dr. du Toit will present to growers and seed company representatives at Texas Spinach Field Day, and Dr. du Toit will present results to American Seed Trade Assoc. Vegetable Tech. Subcommittee. An extension bulletin on Stemphylium leaf spot of spinach will be written and submitted to WSU Extension from Feb. to Apr. 2023. From Mar. to Aug. 2023, a manuscript will be written and submitted by August 2023 on the population genetics of seedborne S. vesicarium (submit to Plant Disease). A final report will be written and submitted for TX Wintergarden Spinach Producers' Board funding part of this project, from Apr. to May 2023. From May to Jun. 2023, a report will be written a report for spinach seed companies. In Jul. 2023, Dr. du Toit will present at the WSU Mount Vernon NWREC Field Day. Kayla will present a poster at the APS conference in Aug. 2023.
Cooperators
- - Producer
- - Producer
- - Producer
- - Producer
Research
Objective 1: Quantify the relative abundance of Stemphylium species pathogenic on spinach that occur on spinach seed lots.
Two spinach seed lots were acquired from each of Denmark (DK1 and 2), France (FR1 and 2), the Netherlands (NL1 and 2), New Zealand (NZ1 and 2), and the United States (USA1 and 2). The seed lots from Denmark, France, the Netherlands, and New Zealand were each harvested in 2017. Two seed lots from the United States were harvested from seed crops in northwestern Washington in 2020 and 2021.
A sample of 100 to 400 seeds of each lot was surface-sterilized with 1.2% NaOCl for 60 s, triple-rinsed in sterilized water, dried, and placed on NP-10 agar medium (Sorensen et al. 1991) to acquire isolates of Stemphylium, as described in the International Seed Health Initiative (ISHI) protocol (ISF 2017). Seeds were incubated at 24°C with a 12 h/12 h day/night cycle, with near-ultraviolet light and cool white fluorescent light by day. Seeds were inspected for the development of pseudothecia or conidia typical of Stemphylium spp., using a dissecting microscope (up to 100x magnification). Individual conidia were isolated by streaking conidia from the seed onto plates of water agar amended with 100 ppm of chloramphenicol (CWA), excising a single conidium from the streaked culture, and placing the conidium onto a plate of clarified V8 agar medium (CV8) on which six sterilized filter disks had been placed. The CV8 agar medium was prepared by centrifuging 200 ml of V8 juice and 4.5 g calcium carbonate at 3,500 rpm for 15 min, and autoclaving 200 ml of the clarified V8 juice with 15 g Bacto agar and 800 ml of deionized water at 15 psi and 121°C for 30 min. If pseudothecia only were observed on a seed, a plug of colonized agar medium was excised from the leading edge of growth and placed onto CV8 medium in a petri plate. Cultures were incubated at room temperature (22 ± 1°C) near a north-facing window to promote sporulation. A single-spore isolate of Stemphylium was then generated from each culture. When the filter disks were fully colonized with mycelium for each single-spore isolate, the disks were placed in a sterilized, paper envelope, dried in a laminar flow hood overnight, and stored in an airtight container with DryRite at room temperature for long-term storage. Isolates of S. vesicarium from the 10 seed lots were used for Objective 2.
In addition, isolates of S. vesicarium were obtained from symptomatic foliage from each of two baby leaf spinach crops (TXfoliar1 and 2). The isolates were obtained from symptomatic leaves collected from each of the two crops grown in the Texas Wintergarden region (near La Pryor, TX) in spring of 2020. Leaves with symptoms typical of Stemphylium leaf spot were collected from each of 50 rows in each crop, labeled, and pressed between blotters in a paper press for medium-term storage. One lesion from each leaf was excised with a sterile scalpel, surface-sterilized in 0.6% NaOCl for 90 s, triple-rinsed in sterilized, deionized water, dried in a laminar flow hood for ~15 min, and placed on CV8 agar medium amended with 100 ppm of chloramphenicol (CCV8). The leaf sections were incubated at room temperature under ambient light near a north-facing window. Isolates of S. vesicarium from the two sets of leaves were used in Objective 3.
A sample of remnant seed of the seed lot used to plant each of the two spinach crops was obtained from the growers (TXseed 1 and 2). One of the seed lots (TXseed1) was treated with fungicides, so the seeds were agitated under running cold tap water for at least 15 min before surface-sterilization to rinse as much of the fungicide treatment off the seed before incubating the seed on NP-10 agar medium. Isolates of S. vesicarium from the two seed lots were used in Objective 3.
To identify Stemphylium isolates from seed and leaves to species, and to select isolates of S. vesicarium for Objectives 2 and 3, each isolate was grown on PDA in the dark to generate aerial mycelium. The mycelium collected for each isolate was stored in a 2-ml tube at -80°C until DNA could be extracted. A FastDNA spin (MP Biomedicals, Irvine, CA) or Synergy Plant (OPS Diagnostics, Lebanon, NJ) DNA extraction kit was used to extract DNA from each isolate using the manufacturer’s protocol, with slight modifications. The DNA was used as a template in a PCR assay to amplify the partial cmdA locus of each isolate, in order to identify each isolate to species using the PCR conditions and phylogeny described by Woudenberg et al. (2017). Amplified DNA of each isolate was sent to ELIM Biopharmaceuticals (Hayward, CA) for Sanger DNA sequencing. Consensus DNA sequences were generated and compared to the NCBI GenBank database through the BLAST algorithm to identify each isolate to species. The incidence of seed on which Stemphylium species were observed was calculated for each seed lot.
A subset of six isolates of S. vesicarium from each of the 14 total populations was tested for pathogenicity to spinach. Subsequently, additional isolates from each population were tested for pathogenicity, for a cumulative 20 to 40 isolates of S. vesicarium from each population as a more accurate indication of the proportion of isolates in each population that are pathogenic to spinach, and how that relates to genetic diversity. Stemphylium species are ubiquitous decomposers of plant debris that can be associated with plants, especially seed (Hernandez-Perez and du Toit 2006), i.e., not all isolates of this genus are plant pathogens, so it is important assess the proportion of isolates in each population that are pathogenic to spinach. Inoculum of each isolate was produced on CV8 agar medium near a north-facing window at room temperature for three weeks to favor sporulation. Spores were dislodged from the agar medium with a sterilized glass rod into ~5 ml of sterilized, distilled water added to each Petri plate, and the suspension strained through two layers of sterilized cheesecloth to remove mycelium. Approximately 24 h before inoculation, three or four replicate spinach plants (~4 weeks old) were enclosed in a large plastic bag per isolate in the greenhouse, and the bags covered with Remay to create a conducive environment for infection and to prevent the plants overheating. Plants were removed from the bags after 24 h, and a spore suspension was sprayed onto the upper and lower leaf surfaces using an atomizer. The plants were then enclosed in the plastic bags again and covered with Remay until the following morning, when the bags were opened. The plants were removed from the bags in the afternoon and placed on a greenhouse bench in a randomized complete block design (RCBD). The greenhouse was maintained at 22 to 25°C with supplemental lighting for 10 h/day. Isolates were determined to be pathogenic to spinach if leaf spots typical of those caused by S. vesicarium were observed 7 days after inoculation.
Objective 2: Assess the role of global spinach seed trade in moving isolates of S. vesicarium, causal agent of Stemphylium leaf spot of spinach, by characterizing populations of S. vesicarium genetically from seed lots grown in the five major spinach seed-producing countries of the world.
Forty isolates of S. vesicarium were obtained from each of the two spinach seed lots originating from each of Denmark, France, the Netherlands, New Zealand, and the USA. The 40 isolates from each of the 10 lots (400 isolates of S. vesicarium) represented 10 populations of S. vesicarium to be evaluated for genetic similarities.
Nine simple sequence repeat (SSR) markers designed originally to characterize S. vesicarium populations that cause Stemphylium leaf blight of onion (Heck et al. 2023) were used to characterize each isolate of S. vesicarium genetically. The markers were tagged with fluorescent dyes and used in two multiplex PCR reactions with DNA extracts of each isolate of S. vesicarium. Amplified DNA was sent to the Cornell University Institute of Biotechnology for fragment analysis on an Applied Biosystems 3730xl DNA Analyzer by capillary electrophoresis. Allele size for amplified DNA of each of the nine markers for each isolate of S. vesicarium was determined manually with the Microsatellite plug-in of the software Geneious Prime. Amplicons representative of each allele for each marker was sent out for Sanger DNA sequencing to confirm the repeat regions.
The R package poppr v.2.9.4 (Kamvar et al. 2014) was used to estimate basic population diversity statistics for each of the 10 seed populations of S. vesicarium. Statistics included the number of multilocus genotypes, expected MLGs (eMLGs), clonal fraction (CF), and Simpson’s diversity index (1 - λ) (Simpson 1949). The signficance of 1 - λ was determined by calculating the 95% confidence interval after 1,000 bootstraps.
Multiple methods were used to examine the underlying population structure of the worldwide S. vesicarium isolates from 10 seed lots. First, clone-corrected data for the isolates were used in a discriminant analysis of principal components (DAPC) (Jombart et al. 2010), calculated using the R package adegenet v.2.1.10 (Jombart 2008), to visualize the structure of the 10 populations. Second, poppr was used to create a minimum spanning network of Bruvo’s genetic distance (Bruvo et al. 2004) for the non-clone-corrected data to determine if genetically similar MLGs are associated with specific seed lots or countries of production.
To test whether the S. vesicarium populations from the 10 seed lots were differentiated regionally (i.e., among countries of production) or locally (i.e., among seed lots), the R package arlequin v.3.5.2.2 (Excoffier and Lischer 2010) was used for Analysis of Molecular Variance (AMOVA) of the data from clone-corrected populations. Country (Denmark, France, the Netherlands, New Zealand, or the United States) of production was used as a grouping factor, with the 10 seed lots nested in their respective countries. A non-parametric test of 1,000 permutations was used to determine the P-value associated with Φ, an analog of FST that estimates genetic differentiation based on covariance components (Excoffier et al. 1992) for each hierarchical level of the AMOVA.
To determine whether pathogenicity of isolates to spinach impacted the population structure of the worldwide seed populations, a similar approach to that described above was taken using the subset of 248 isolates of S. vesicarium that were tested for pathogenicity. First, isolates were stratified into two groups, pathogenic or non-pathogenic to spinach, with subpopulations being the 10 seed lots. The non-clone-corrected data were used to calculate the same summary statistics described above. The data were then clone-corrected based on each of the 10 seed lots in each of pathogenic and non-pathogenic groups. A DAPC of the pathogenic and non-pathogenic groups as subpopulations within each of the 10 seed lots, for a total of 15 groups, was estimated to determine if populations were structured based on the seed lot from which the pathogenic or non-pathogenic isolates originated. A minimum spanning network of Bruvo’s distance of the 15 groups was generated for the non-clone-corrected data to evaluate whether the genetic relatedness of MLGs was associated with pathogenic and non-pathogenic isolates among the 10 seed lots. A hierarchical AMOVA was calculated to determine if the populations of S. vesicarium differed based on pathogenicity to spinach, or seed lot.
Objective 3: Determine the relative significance of seedborne S. vesicarium as an inoculum source for outbreaks of Stemphylium leaf spot by comparing isolates of the fungus from leaves of baby leaf spinach crops symptomatic with the disease to isolates from seed lots used to plant these spinach crops.
Fifty isolates of S. vesicarium were collected from each of two fresh market spinach crops in Texas in 2020 that had developed symptoms of Stemphylium leaf spot. In addition, 40 isolates of S. vesicarium were collected from a sample of the seed lot used to plant each crop. The 50 isolates of S. vesicarium from leaves of each of the two spinach crops and 40 isolates from each of the two seed lots planted represent four populations to be assessed for Objective 3. The same analyses as those described above were used. Only the 110 isolates that were tested for pathogenicity were used in analyses because of the observed impact of pathogenicity on population structure in Objective 2.
Objective 1: Quantify the relative abundance of Stemphylium species pathogenic on spinach that occur on spinach seed lots.
The total incidence of Stemphylium on the 12 seed lots seed ranged from 2.5 to 73.5% (Table 1). S. vesicarium was the predominant species, with the incidence on seed ranging from 1.1 to 70.6%. This species made up 42.7 to 100% of the Stemphylium isolates obtained from each seed lot. The next most predominant species was S. beticola, at an incidence ranging from 0.2 to 5.9% of seed. This species made up 2.3 to 56.3% of the Stemphylium isolates obtained from each seed lot. Other species that were isolated from fewer seed per lot included S. eturmiunum, S. amaranthi, S. astragali, S. simmonsii, S. chrysanthemicola, S. lucomagnoense, S. gracilariae, and S. drummondii. The only species documented to be pathogenic to spinach are S. vesicarium, S. beticola, and S. drummondii (Liu et al. 2020; Spawton et al. 2019; Zhou et al. 2011). A subset of isolates of each species from these seed lots, besides those of S. vesicarium tested, was tested for pathogenicity on the spinach cv. Mandolin, including S. beticola (n = 7), S. eturmiunum (4), S. amaranthi (4), S. astragali (1), S. simmonsii (5), S. chrysanthemicola (1), S. lucomagnoense (1), S. gracilariae (1), and S. drummondii (1). None of these isolates caused leaf spots on spinach, even though S. beticola and S. drummondii are known spinach pathogens. The lack of pathogenicity of isolates of the latter two species may reflect loss of virulence when isolates are stored for an extended period, as has been documented by other researchers working with Stemphylium species that are pathogenic on spinach (Stephen Koike, TriCal Diagnostics).
Table 1. Incidence (%) of Stemphylium species isolated from 12 spinach seed lots
|
Seed lot |
Year of production |
# of seed plated |
Incidence (%) of seed (and percentage of Stemphylium isolates of that species on that lot)a |
||||||||||
|
S. vesi-carium |
S. beticola |
S. eturm-iunum |
S. amar-anthi |
S. astra-gali |
S. simmo-nsii |
S. chrysan-themicola |
S. lucomag-noense |
S. gracil-ariae |
S. drumm-ondii |
Total incidence |
|||
|
DK1 |
2017 |
678 |
6.2 (89.4) |
0.3 (4.3) |
-b |
0.1 (2.1) |
0.1 (2.1) |
|
|
0.1 (2.1) |
- |
- |
6.9 |
|
DK2 |
2017 |
542 |
7.6 (93.2) |
0.2 (2.3) |
- |
0.2 (2.3) |
0.2 (2.3) |
- |
- |
- |
- |
8.1 |
|
|
FR1 |
2017 |
172 |
30.8 (100) |
- |
- |
- |
- |
- |
- |
- |
- |
- |
30.8 |
|
FR2 |
2017 |
610 |
6.9 (82.4) |
0.8 (9.8) |
0.3 (3.9) |
- |
0.3 (3.9) |
- |
- |
- |
- |
- |
8.4 |
|
NL1 |
2017 |
508 |
8.9 (86.5) |
- |
0.4 (3.8) |
0.6 (5.8) |
0.2 (1.9) |
- |
0.2 (1.9) |
- |
- |
- |
10.2 |
|
NL2 |
2017 |
508 |
8.3 (100) |
- |
- |
- |
- |
- |
- |
- |
- |
- |
8.3 |
|
NZ1 |
2017 |
576 |
9.7 (54.9) |
5.9 (33.3) |
1.6 (8.8) |
0.2 (1.0) |
- |
- |
- |
0.2 (1.0) |
0.2 (1.0) |
- |
17.7 |
|
NZ2 |
2017 |
576 |
8.2 (45.2) |
5.9 (32.7) |
3.8 (21.2) |
- |
0.2 (1.0) |
- |
- |
- |
- |
- |
18.1 |
|
USA1 |
2020 |
68 |
70.6 (96.0) |
2.9 (4.0) |
- |
- |
- |
- |
- |
- |
- |
- |
73.5 |
|
USA2 |
2021 |
170 |
28.2 (88.9) |
1.8 (5.6) |
0.6 (1.9) |
- |
- |
0.6 (1.9) |
- |
- |
- |
0.6 (1.9) |
31.8 |
|
TXseed1c,d |
Unknown |
3,806 |
1.1 (42.7) |
1.4 (56.3) |
- |
<0.1 (1.0) |
- |
- |
- |
- |
- |
- |
2.5 |
|
TXseed2d |
Unknown |
2,546 |
1.9 (54.4) |
0.6 (17.8) |
<0.1 (1.1) |
<0.1 (1.1) |
0.2 (6.7) |
0.3 (7.8) |
- |
<0.1 (1.1) |
0.4 (10.0) |
- |
3.5 |
a Seed of each lot was surface-sterilized in 1.2% NaOCl for 90 s, triple-rinsed in sterile, de-ionized water, dried, plated on NP-10 agar medium (Sorensen et al. 1991), and incubated as described by the International Seed Federation (2017). Single-spore isolations were carried out as described in the main text. Species determination was based on the best match of the cmdA sequence of the isolate to cmdA accessions in GenBank. All sequences were 95.29% identical to the best matching GenBank sequences of the species listed. The number in parentheses is the percentage of isolates identified as that species out of all Stemphylium isolates from that seed lot.
b - = the species was not isolated from the seed lot.
c Seed lot planted for a spinach crop in Texas in 2020 that developed symptoms of Stemphylium leaf spot.
d Seed lot was treated with a fungicide, so seed was rinsed under running deionized water for 15 min before sterilization and incubation on NP-10 agar medium for isolations.
The percentage of S. vesicarium isolates pathogenic to spinach from each of the 10 seed populations used in Objective 2 ranged from 0 to 100% (Table 2). Many vegetable seed companies select which spinach seed lots to treat with hot water for certified organic control of seedborne Stemphylium spp. based on the incidence of seed on which Stemphylium spp. are observed. The presence of significant proportions of non-pathogenic isolates on seed lots in this study suggests that companies may overestimate the amount of pathogenic S. vesicarium.
Table 2. Summary population statistics for non-clone corrected data for populations of Stemphylium vesicarium from 10 spinach seed lots used in a worldwide seed population study, and a subset of isolates tested for pathogenicity to spinach, with isolates genotyped using nine simple sequence (SSR) markers
|
Analysis by |
Year |
Location |
Na |
MLGsb |
eMLGsc |
Unique MLGsd |
CFe |
1 - λf |
# of isolates tested for pathogenicity (% pathogenic)g |
|
|
Seed loth |
||||||||||
|
DK1 |
2017 |
Denmark |
40 |
37 |
37.00 |
31 |
0.075 |
0.971 (0.959-0.984) |
29 (3.4) |
|
|
DK2 |
2017 |
Denmark |
40 |
14 |
14.00 |
7 |
0.650 |
0.623 (0.455-0.790) |
25 (72.0) |
|
|
FR1 |
2017 |
France |
40 |
5 |
5.00 |
1 |
0.875 |
0.230 (0.056-0.404) |
20 (100.0) |
|
|
FR2 |
2017 |
France |
40 |
39 |
39.00 |
36 |
0.025 |
0.974 (0.963-0.985) |
26 (3.8) |
|
|
NL1 |
2017 |
Netherlands |
40 |
38 |
38.00 |
35 |
0.050 |
0.973 (0.960-0.985) |
29 (0.0) |
|
|
NL2 |
2017 |
Netherlands |
40 |
27 |
27.00 |
24 |
0.325 |
0.941 (0.912-0.970) |
29 (0.0) |
|
|
NZ1 |
2017 |
New Zealand |
40 |
22 |
22.00 |
15 |
0.450 |
0.901 (0850-0.953) |
24 (29.2) |
|
|
NZ2 |
2017 |
New Zealand |
40 |
35 |
35.00 |
33 |
0.125 |
0.961 (0.939-0.984) |
26 (0.0) |
|
|
USA1 |
2020 |
WA, USA |
40 |
2 |
2.00 |
0 |
0.950 |
0.219 (0.065-0.372) |
20 (100.0) |
|
|
USA2 |
2021 |
WA, USA |
40 |
7 |
7.00 |
2 |
0.825 |
0.614 (0.527-0.700) |
20 (85.0) |
|
|
Total |
400 |
199 |
184 |
|||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
Pathogenicityi |
||||||||||
|
Pathogenic |
84 |
11 |
11.00 |
11 |
0.869 |
0.628 (0.529-0.726) |
||||
|
DK1 |
2017 |
Denmark |
1 |
1 |
1.00 |
0 |
||||
|
DK2 |
2017 |
Denmark |
18 |
7 |
4.75 |
2 |
||||
|
FR1 |
2017 |
France |
20 |
5 |
3.26 |
1 |
||||
|
FR2 |
2017 |
France |
1 |
1 |
1.00 |
1 |
||||
|
NZ1 |
2017 |
New Zealand |
7 |
5 |
5.00 |
1 |
||||
|
USA1 |
2020 |
WA, USA |
20 |
2 |
1.98 |
0 |
||||
|
USA2 |
2021 |
WA, USA |
17 |
4 |
3.18 |
0 |
||||
|
Non-pathogenic |
164 |
158 |
82.30 |
158 |
0.037 |
0.993 (0.992-0.995) |
||||
|
DK1 |
2020 |
WA, USA |
28 |
28 |
10.00 |
26 |
||||
|
DK2 |
2021 |
WA, USA |
7 |
7 |
7.00 |
6 |
||||
|
FR2 |
2017 |
France |
25 |
25 |
10.00 |
24 |
||||
|
NL1 |
2017 |
Netherlands |
29 |
29 |
10.00 |
29 |
||||
|
NL2 |
2017 |
Netherlands |
29 |
27 |
9.78 |
26 |
||||
|
NZ1 |
2017 |
New Zealand |
17 |
17 |
10.00 |
16 |
||||
|
NZ2 |
2017 |
New Zealand |
26 |
26 |
10.00 |
26 |
||||
|
USA2 |
2021 |
WA, USA |
3 |
3 |
3.00 |
2 |
||||
a Number of isolates genotyped in each population.
b Number of multilocus genotypes (MLGs) in that population. This is also the number of individuals in each population after clone-correction.
c Expected number of MLGs after rarefaction at the smallest sample size.
d Number of MLGs found only in that population.
e Clonal fraction, estimated as 1 - (MLG/N).
f Simpson’s diversity index (Simpson 1949). Numbers in parentheses is the 95% confidence interval after 1,000 bootstraps.
g At least 50% of the isolates from each seed lot were tested for pathogenicity to the spinach cv. Mandolin.
h Summary statistics calculated for the population of S. vesicarium from each of the 10 seed lots
i Summary statistics calculated for the isolates from each seed lot tested for pathogenicity, with each tested isolate categorized as pathogenic or non-pathogenic. The number of isolates from each seed lot that comprised the pathogenic and non-pathogenic groups is displayed.
Of the 110 isolates of S. vesicarium for Objective 3 that were tested for pathogenicity, 56 were pathogenic to spinach and present in three of the four populations: TXfoliar1, TXfoliar2, and TXseed1 (Table 3). In contrast, the 54 isolates that did not cause Stemphylium leaf spot were only present in the seed populations, i.e., all the foliar isolates were pathogenic to spinach.
Table 3. Summary statistics of non-clone corrected data for four spinach populations of Stemphylium vesicarium for a seed vs. foliar population study, and for the subset of isolates tested for pathogenicity, with isolates genotyped using nine simple sequence (SSR) markers
|
Analysis by |
Tissue |
Year |
Location |
Na |
MLGsb |
eMLGsc |
Unique MLGsd |
CFe |
1 - λf |
# of isolates tested for pathogenicity (% pathogenic)g |
|
|
Populationh |
|||||||||||
|
TXfoliar1 |
Leaf |
2020 |
TX, USA |
50 |
6 |
5.56 |
2 |
0.880 |
0.536 (0.406-0.666) |
25 (100.0) |
|
|
TXseed1 |
Seed |
Unknown |
Denmark |
40 |
30 |
30.00 |
27 |
0.250 |
0.959 (0.941-0.977) |
31 (19.4) |
|
|
TXfoliar2 |
Leaf |
2020 |
TX, USA |
50 |
5 |
4.39 |
1 |
0.900 |
0.221 (0.067-0.375) |
25 (100.0) |
|
|
TXseed2 |
Seed |
Unknown |
Unknown |
40 |
35 |
35.00 |
34 |
0.125 |
0.968 (0.952-0.983) |
29 (0.0) |
|
|
Total |
180 |
68 |
64 |
||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Pathogenicityi |
|||||||||||
|
Pathogenic |
56 |
11 |
|||||||||
|
TXfoliar1 |
Leaf |
2020 |
TX, USA |
25 |
6 |
4.20 |
2 |
||||
|
TXfoliar2 |
Leaf |
2020 |
TX, USA |
25 |
5 |
3.00 |
1 |
||||
|
TXseed1 |
Seed |
Unknown |
Denmark |
6 |
6 |
6.00 |
3 |
||||
|
Non-pathogenic |
|
54 |
52 |
||||||||
|
TXseed1 |
Seed |
Unknown |
Denmark |
25 |
25 |
25.00 |
24 |
||||
|
TXseed2 |
Seed |
Unknown |
Unknown |
29 |
28 |
24.30 |
27 |
||||
a Number of isolates genotyped in each population.
b Number of multilocus genotypes (MLGs) in that population. This is also the number of individuals in each population after clone-correction.
c Expected number of MLGs after rarefaction at the smallest sample size.
d Number of MLGs found only in that population.
e Clonal fraction, calculated as 1 - (MLG/N).
f Simpson’s diversity index (Simpson 1949). Numbers in parentheses is the 95% confidence interval after 1,000 bootstraps.
g At least 50% of the isolates from each of the four populations were tested for pathogenicity to the spinach cv. Mandolin.
h Summary statistics calculated for the populations of S. vesicarium from each of the four populations.
i Summary statistics calculated for isolates from each population tested for pathogenicity, with each tested isolate categorized as pathogenic or non-pathogenic. The number of isolates from each population that comprised the pathogenic and non-pathogenic groups is displayed.
Objective 2: Assess the role of global spinach seed trade in moving isolates of S. vesicarium, causal agent of Stemphylium leaf spot of spinach, by characterizing populations of S. vesicarium genetically from seed lots grown in the five major spinach seed-producing countries of the world.
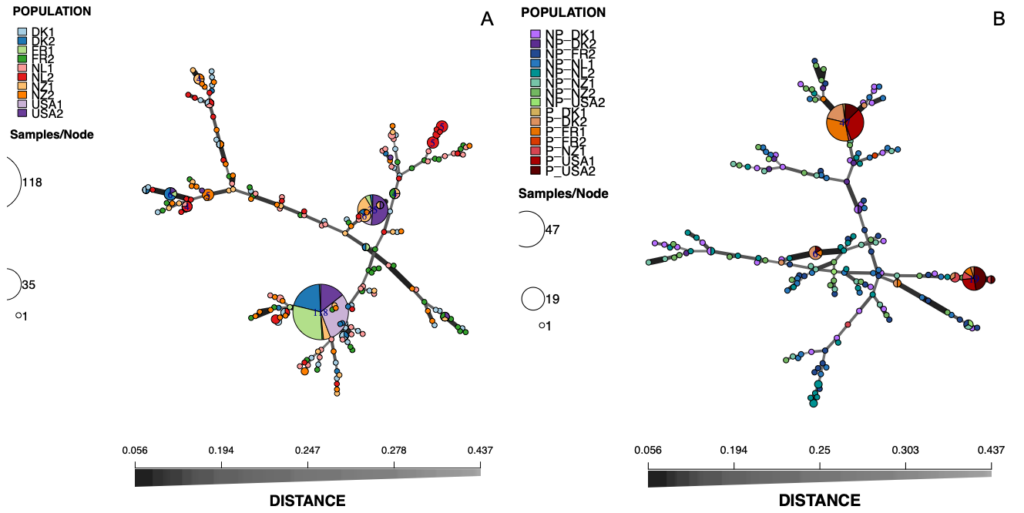
In total, 199 multilocus genotypes (MLGs) were detected in the 400 S. vesicarium isolates from spinach seed, ranging from 2 for seed lot USA1 to 39 for lot FR2 (Table 2). The number of MLGs unique to each population (i.e., only occurring in one of the 10 populations) ranged from 0 for lot USA1 to 36 for FR2, with 184 of the 199 total MLGs unique to specific seed lots. Diversity, based on Simpson’s diversity index (1 - λ), ranged from 0.219 for USA1 to 0.974 for FR2. The most frequent MLG was MLG.167, which was detected in 118 of the 400 isolates, in 7 of the 10 seed populations (1 isolate from DK1, 24 from DK2, 35 from FR1, 1 from FR2, 5 from NZ1, 35 from USA1, and 17 from USA2; Fig. 1A). The next most frequent MLG was MLG.115, detected in 35 of the isolates from five of the seed populations (1 isolate from DK2, 2 from FR1, 9 from NZ1, 5 from USA1, and 18 from USA2). Of the 199 MLGs detected, 175 (88%) were comprised of only one isolate.
Of the subset of 248 seed isolates of S. vesicarium that were tested for pathogenicity, the 84 that were pathogenic to the spinach cv. Mandolin originated from 7 of the 10 seed lots: DK1, DK2, FR1, FR2, NZ1, USA1, and USA2 (Table 2). The 164 isolates that did not cause Stemphylium leaf spot originated from 8 seed lots: DK1, DK2, FR2, NL1, NL2, NZ1, NZ2, and USA2. Therefore, 169 MLGs were detected in the 248 isolates tested for pathogenicity to spinach, of which 158 MLGs (93%) were each comprised of a single isolate. The 84 pathogenic isolates belonged to 11 MLGs, all of which were unique to the pathogenic isolates, while the 164 non-pathogenic isolates belonged to 158 MLGs (82.30 eMLGs), with all 158 MLGs unique to the non-pathogenic isolates. Diversity, measured as the Simpson’s diversity index (1 - λ), was greater for the non-pathogenic isolates (0.993) than the pathogenic isolates (0.628). Therefore, there appears to be an inverse correlation between pathogenicity to spinach and genetic diversity, i.e., populations of S. vesicarium on seed comprised mostly of pathogenic isolates tended to be less diverse genetically. MLG.167, the most frequent MLG detected in the original 400 S. vesicarium seed isolates, was also the most frequent MLG in the 248 isolates tested for pathogenicity, present in 47 of the isolates from six seed lots: 1 isolate from DK1, 9 from DK2, 15 from FR1, 1 from NZ1, 15 from USA1, and 6 from USA2, with all of these isolates pathogenic to spinach (Fig. 1B). Similarly, MLG.155, the second most frequent MLG among the 400 seedborne isolates, was also the second most frequent MLG detected in the 248 isolates tested for pathogenicity, and occurred in 19 of the isolates from five seed lots: 1 from DK2, 2 from FR1, 2 from NZ1, 5 from USA1, and 9 from USA2, all of which were pathogenic to spinach (Fig. 1B).
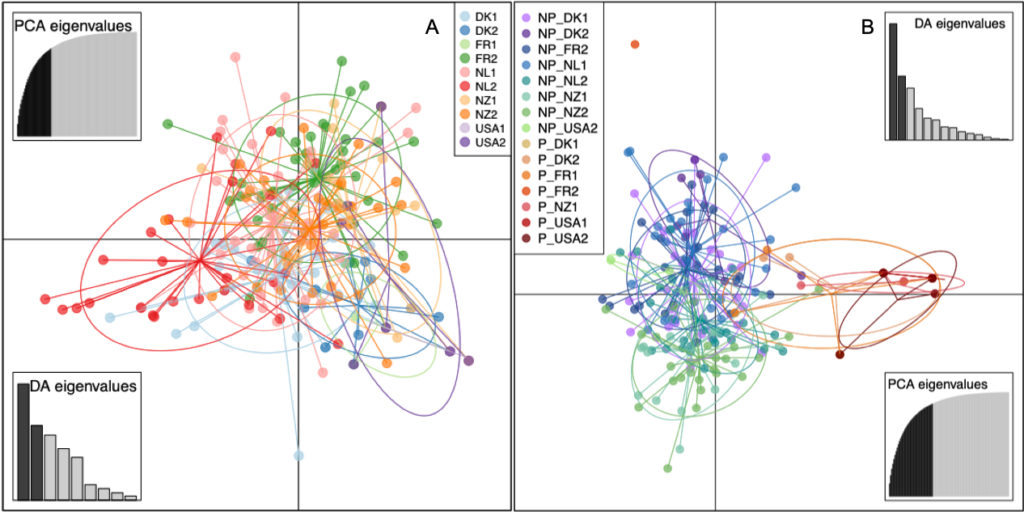
The DAPC of the 40 isolates of S. vesicarium isolates from each of the 10 spinach seed lots, did not differentiate the 10 seed populations by seed lot or country of origin (Fig. 2A). Similarly, the minimum spanning network of Bruvo’s distance for the non-clone corrected data of S. vesicarium from the 10 seed lots illustrated no clear clustering of MLGs based on seed lot, as the MLGs making up each seed lot were dispersed across the network (Fig. 1A). The AMOVA corroborated results of the DAPC and minimum spanning network. When the country in which the seed lot was grown was used as a grouping factor for the clone-corrected data of the S. vesicarium isolates from 10 seed lots, a large proportion of the genetic variation existed within seed lots (97.74%, P = 0.001; Table 4). The effect of the country in which the seed lot was grown was not significant (1.22%, P = 0.122). The effect of the seed lot within country was significant at P = 0.055, but only comprised 1.04% of the genetic variation. Therefore, the AMOVA indicated there was insignificant local or regional structure of this worldwide population of seedborne isolates of S. vesicarium. This could be due to unrestricted gene flow among the major countries in which spinach seed is produced, likely as a result of frequent movement of seed that limits regional and local genetic differentiation of populations of S. vesicarium. Given the nature of spinach seed production, whereby stock seed produced in one location typically is moved to various countries where seed crops are produced, and the harvested seed is then moved to even more countries where processing and fresh market spinach are grown, it is not surprising there was no evidence of genetic differentiation of seedborne populations of S. vesicarium based on the country of seed production. It is also possible that a lack of genetic differentiation is indicative of spinach being a new host for S. vesicarium, as this disease was only first described in 2001 (Koike et al. 2001).
Table 4. Analysis of molecular variance (AMOVA) of clone corrected data for populations of Stemphylium vesicarium from 10 spinach seed lots using a worldwide seed population study, in which isolates were genotyped with nine simple sequence repeat (SSR) markers, with geographic location used as a grouping factor
|
Source of variation |
Df |
Sum of squares |
Variance component (𝜎) |
Variation (%) |
Φa |
Pb |
|
Among countries |
4 |
46.14 |
0.079 |
1.22 |
0.012 |
0.122 |
|
Among seed lots within countries |
5 |
37.94 |
0.068 |
1.04 |
0.011 |
0.055 |
|
Within seed lots |
216 |
1,373.09 |
6.357 |
97.74 |
0.023 |
0.001 |
|
Total |
225 |
1,457.17 |
6.504 |
100.00 |
a An analog of FST that estimates genetic differentiation based on covariance components (Excoffier et al. 1992). A larger value suggests greater genetic differentiation.
b Significance of Φ was tested using a randomization test of 999 permutations in the R package arlequin (Excoffier and Lischer 2010).
When a second DAPC was conducted in which clone-corrected S. vesicarium isolates were organized by the 15 groups of pathogenic or non-pathogenic isolates within seed lots, the first DAPC axis separated most of the pathogenic isolates within seed lots (from DK1, DK2, FR1, NZ1, USA1, and USA2) from the non-pathogenic isolates, although there was some overlap between the pathogenic and non-pathogenic groups (Fig. 2B). The second DAPC axis (vertical axis) separated the one pathogenic isolate in FR2 from the other pathogenic and non-pathogenic isolates from all 10 seed lots. The minimum spanning network of Bruvo’s distance for the non-clone-corrected populations indicated that the three MLGs that were most frequent in the pathogenic isolates within each seed lot were more closely related to non-pathogenic MLGs than to one another, as these pathogenic MLGs were distributed across the network (Fig. 1B). The pathogenic isolates comprising these three major MLGs were similar genetically to several pathogenic isolates comprising one or two other MLGs, suggesting that some pathogenic isolates may be closely related, but not all pathogenic isolates are closely related. Additionally, there was no clear clustering of the MLGs detected in non-pathogenic isolates from the same seed lot (Fig. 1B), similar to what was demonstrated in the first DAPC with the data set for the original 400 seedborne isolates of S. vesicarium from the 10 seed lots (Fig. 2A).
When pathogen vs. non-pathogen was used as a grouping factor in the AMOVA, most of the variation occurred within seed lots (89.73%, P = 0.001; Table 5). The effect of pathogen vs. non-pathogen was also significant (9.16%, P = 0.001), as was the effect of seed lot within pathogenic and non-pathogenic groups (1.11%, P = 0.006), but to a lesser degree than the variation within seed lots. This suggests that pathogenic vs. non-pathogenic status of isolates does impact the genetic variation among worldwide populations of S. vesicarium on spinach.
Table 5. Analysis of molecular variance (AMOVA) of clone corrected data for populations of Stemphylium vesicarium from 10 spinach seed lots for a worldwide seed population study using isolates tested for pathogenicity to spinach, with isolates genotyped using nine simple sequence repeat (SSR) markers, and pathogenic vs. non-pathogenic as a grouping factor
|
Source of variation |
Df |
Sum of squares |
Variance component (𝜎) |
Variation (%) |
Φa |
Pb |
|
Pathogenic vs. non-pathogenic populations |
1 |
34.59 |
0.640 |
9.16 |
0.092 |
0.001 |
|
Among seed lots within pathogenic or non- pathogenic populations |
13 |
93.76 |
0.078 |
1.11 |
0.012 |
0.006 |
|
Within seed lots |
172 |
1,078.62 |
6.271 |
89.73 |
0.103 |
0.001 |
|
Total |
186 |
1,206.97 |
6.989 |
100.00 |
a An analog of FST that estimates genetic differentiation based on covariance components (Excoffier et al. 1992). A larger value suggests greater genetic differentiation.
b Significance of Φ was tested using a randomization test of 999 permutations in the R package arlequin (Excoffier and Lischer 2010).
Objective 3: Determine the relative significance of seedborne S. vesicarium as an inoculum source for outbreaks of Stemphylium leaf spot by comparing isolates of the fungus from leaves of baby leaf spinach crops symptomatic with the disease to isolates from seed lots used to plant these spinach crops.
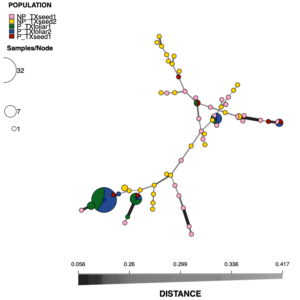
Sixty-eight MLGs were observed among the 180 isolates of S. vesicarium (Table 3). The foliar populations had 6 (TXfoliar1) and 5 (TXfoliar2) MLGs, while the two seed populations had 30 (TXseed1) and 35 (TXseed2) MLGs. The number of MLGs unique to each population was far less for the foliar populations (2 for TXfoliar1 and 1 for TXfoliar2) compared to the seed populations (27 for TXseed1 and 34 for TXseed2). Diversity, based on the Simpson’s diversity index (1 - λ) was 0.536 and 0.221 for TXfoliar1 and TXfoliar2 populations, respectively, and was much greater for the seed populations at 0.959 and 0.968 for TXseed1 and TXseed2, respectively. The most frequent MLG was MLG.68, found in 76 isolates, including 32 isolates from TXfoliar1 and 44 from TXfoliar2. The next most frequent MLG was MLG.167, which was also the most frequent MLG detected in the worldwide seed population study. MLG.167 was found in 13 isolates from three populations in this foliar vs. seed population study (11 isolates from TXfoliar1, 1 from TXfoliar2, and 1 from TXseed1). The prevalence of MLG.167 in both the worldwide seed populations and the foliar populations of S. vesicarium suggests that this MLG may be common in both seed and foliar populations beyond the populations evaluated in these studies. Of the 68 MLGs, 53 (78%) were each detected in only one isolate, with all but one of the 53 MLGs detected in one of the two S. vesicarium populations from seed, exemplifying the high diversity of the seed populations compared to the foliar populations of S. vesicarium.
The pathogenic isolates belonged to 11 MLGs, while the 54 non-pathogenic isolates belonged to 52 MLGs, with only two MLGs shared between the pathogenic and non-pathogenic isolates. MLG.68, the most frequent MLG detected in the original 180 S. vesicarium isolates in the study, was also the most frequent MLG detected in the 110 isolates tested for pathogenicity, occurring in 32 isolates (57% of pathogenic isolates) from the foliar populations, 13 from TXfoliar1 and 19 in TXfoliar2, but not in any of the seed isolates (Fig. 3). Similarly, MLG.167, the second most frequent MLG among the original 180 isolates, was the second most frequent MLG in the 110 isolates tested for pathogenicity, occurring in 7 pathogenic isolates (13% of pathogenic isolates): 5 in TXfoliar1, 1 in TXfoliar2, and 1 in TXseed1.
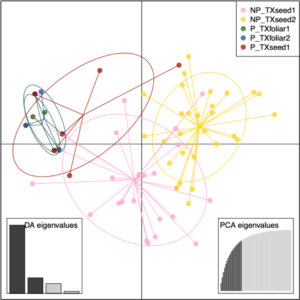
The DAPC of the subset of 110 isolates tested for pathogenicity from the four populations in the seed vs. foliar population study clustered the pathogenic isolates of S. vesicarium together (Fig. 4). The first DAPC axis separated the two pathogenic foliar populations and the single pathogenic seed population (from TXseed1) from both non-pathogenic seed populations, with limited overlap of the pathogenic isolates with non-pathogenic isolates in TXseed1, and limited overlap of the two non-pathogenic populations. The second DAPC axis separated the non-pathogenic TXseed1 population from the other three populations. This suggests that the structure of the two pathogenic foliar populations was most similar, and these foliar populations shared some genetic similarities with the pathogenic isolates from TXseed1, and pathogenic isolates from TXseed1 had some genetic similarities to the non-pathogenic isolates from TXseed1. The non-pathogenic TXseed1 isolates shared some genetic similarities with all four populations.
The minimum spanning network of Bruvo’s distance indicated that the two MLGs frequently detected in the pathogenic isolates (MLG.68 and MLG.167) were closely related, along with four MLGs that had one or two pathogenic isolates each, while the remaining five MLGs present in the pathogenic isolates were distantly related (Fig. 3). Only three MLGs (MLG.23, MLG.32, and MLG.37) were shared by pathogenic isolates from both foliage and seed, and these MLGs were distantly related. These shared MLGs suggests that S. vesicarium isolates that cause Stemphylium leaf spot of spinach can originate from infected seed, as demonstrated previously for seedborne S. beticola (Hernandez-Perez 2005). The fact that few pathogenic isolates from seed were found is not surprising given that very little infected seed is needed for the disease to occur in fresh market crops under optimal conditions of high temperatures and humidity (du Toit and Ocamb 2023; Koike et al. 2001), especially given that baby leaf spinach crops are planted at 7 to 9 million seed/ha, with sequential plantings under sprinkler irrigation. Therefore, Stemphylium leaf spot could occur even at a seed transmission rate of 10-6 seed planted (which would result in >1 infected spinach seedling/ha). Given the high diversity of isolates on seed, there are likely more MLGs than those detected from just 40 isolates per seed lot, some of which may be present in the leaf populations.
Based on the AMOVA to determine if pathogenicity of isolates of S. vesicarium to spinach differentiated the two populations from seed and the two populations from foliage, most of the variation among isolates was detected within each of the four populations (94.22% P = 0.013, Table 6). The effect of pathogenic vs. non-pathogenic groups was also significant (5.80%, P = 0.001), but the effect of population within pathogenic and non-pathogenic groups was not significant (-0.02%, P = 0.350). The significant effect of pathogenicity corroborates results of the DAPC for this study (Fig. 4), as well as results from the worldwide seed population study.
Table 6. Analysis of molecular variance (AMOVA) of clone corrected data for four spinach populations of Stemphylium vesicarium used in a seed vs. foliar population study and tested for pathogenicity to spinach, with isolates genotyped using nine simple sequence repeat (SSR) markers, and pathogenic vs. non-pathogenic groups used as the grouping factor
|
Source of variation |
Df |
Sum of squares |
Variance component (𝜎) |
Variation (%) |
Φa |
Pb |
|
Pathogenic vs. non-pathogenic groups |
1 |
16.03 |
0.382 |
5.80 |
0.058 |
0.001 |
|
Among populations within pathogenic or non- pathogenic populations |
3 |
18.56 |
-0.002 |
-0.02 |
0.000 |
0.350 |
|
Within populations |
65 |
403.49 |
6.208 |
94.22 |
0.058 |
0.013 |
|
Total |
69 |
438.09 |
6.588 |
100.00 |
a An analog of FST that estimates genetic differentiation based on covariance components (Excoffier et al. 1992). A larger value suggests greater genetic differentiation.
b Significance of Φ was tested using a randomization test of 999 permutations in the R package arlequin (Excoffier and Lischer 2010).
References
Bruvo, R., Michiels, N. K., D’Souza, T. G., and Schulenburg, H. 2004. A simple method for calculation of microsatellite genotype distances irrespective of ploidy level. Mol. Ecol. 13:2101-2106.
du Toit, L. J., and Ocamb, C. M. 2023. Spinach (Spinacia oleracea) - Stemphylium leaf spot. In: Pacific Northwest Plant Disease Management Handbook [online]. J. W. Pscheidt and C. M. Ocamb, eds. Oregon State University, Corvallis. https://pnwhandbooks.org/plantdisease/host-disease/spinach-spinacia-oleracea-stemphylium-leaf-spot. Accessed 23 May 2023.
Excoffier, L., and Lischer, H. E. L. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux or Windows. Mol. Ecol. Resourc. 10:564-567.
Excoffier, L., Smouse, P. E., and Quattro, J. M. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genet. 131:479-491.
Heck, D. W., Hay, F., and Pethybridge, S. J. 2023. Enabling population biology studies of Stemphylium vesicarium from onion with microsatellites. Plant Dis In press.
Hernandez-Perez, P. 2005. Seed transmission of Stemphylium botryosum and Cladosporium variabile in spinach. Pp. 79-101, in: Management of seedborne Stemphylium botryosum and Cladosporium variabile causing leaf spot of spinach seed crops in Western Washington. M.S. thesis, Washington State University, Pullman.
Hernandez-Perez, P., and du Toit, L. J. 2006. Seedborne Cladosporium variabile and Stemphylium botryosum in spinach. Plant Dis. 90:137-145.
International Seed Federation. 2017. Methods for the detection of Verticillium dahliae on spinach seed. http://worldseed.org/wp-content/uploads/2017/07/Spinach_Verticillium_July2017.pdf
Jombart, T. 2008. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinform. 24:1403-1405.
Jombart, T., Devillard, S., and Balloux, F. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11:94.
Kamvar, Z. N., Tabima, J. F., and Grünwald, N. J. 2014. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281.
Koike, S. T., Henderson, D. M., and Butler, E. E. 2001. Leaf spot disease of spinach in California caused by Stemphylium botryosum. Plant Dis. 85:126-130.
Liu, B., Stein, L., Cochran, K., du Toit, L. J., Feng, C., Dhillon, B., and Correll, J. C. 2020. Characterization of lead spot pathogens from several spinach production areas in the United States. Plant Dis. 104:1994-2004.
Sorensen, L. H., Schneider, A. T., and Davis, J. R. 1991. Influence of sodium polygalacturonate sources and improved recovery of Verticillium spp. from soil. Phytopathology 81:1347. (Abstr.)
Spawton, K. A., Derie, M. L., Correll, J. C., Stein, L., Olaya, G., Raid, R. N., Sandoya, G. V., Peever, T. L., and du Toit, L. J. 2019. Characterization of Stemphylium spp. from spinach based on molecular data, host response, and azoxystrobin sensitivity. Phytopathology 109:S2.167. Poster presented at Plant Health 2019, Annual Meeting of American Phytopathological Society, 3-7 Aug. 2019, Cleveland, OH.
Woudenberg, J. H. C., Hanse, B., van Leeuwen, G. C. M., Groenewald, J. Z., and Crous, P. W. 2017. Stemphylium revisited. Studies Mycol. 87:77-103.
Zhou, Y., Shi, Y., Xie, X., Guo, Y., and Li, B. 2011. Leaf spot of spinach caused by Stemphylium spinaciae sp. nov. Mycostema 30:379-383.
Research Outcomes
The presence of significant proportions of non-pathogenic isolates on seed lots in this study suggests that companies may overestimate the amount of pathogenic S. vesicarium (or S. beticola) when testing spinach seed lots for the causal agents of Stemphylium leaf spot. The methods used to quantify Stemphylium on spinach seed, including the freeze-blotter assay (du Toit and Hernandez-Perez 2005), NP-10 agar medium assay (ISF 2017), and indirect molecular PCR assays (Feng et al. 2014; Liu et al. 2021), cannot differentiate isolates of S. vesicarium that are pathogenic to spinach from those that are non-pathogenic. Spinach seed lots typically do not show signs of the pathogen on seed until the seed is incubated, and S. vesicarium can infect both the pericarp and embryo of seed (Hernandez-Perez and du Toit 2006). Therefore, there is a need to develop seed testing methods that reflect more accurately the phytosanitary risk of spinach seed harboring pathogenic isolates of S. vesicarium and S. beticola. Development of diagnostic tools with the ability to differentiate pathogenic from non-pathogenic isolates of S. vesicarium should be based on genetic factors associated with pathogenicity to spinach. Köhl et al. (2009) designed a quantitative, real-time PCR assay to differentiate non-pathogenic isolates from pathogenic isolates of S. vesicarium causing brown spot of pear. Designing a similar protocol to differentiate non-pathogenic vs. pathogenic isolates of S. vesicarium associated with spinach would be valuable for assessing more accurately the risk of seedborne inoculum of S. vesicarium causing outbreaks of Stemphylium leaf spot.
Pathogenic isolates of S. vesicarium from foliar populations shared MLGs with pathogenic isolates from one of the two seed populations. This suggests that S. vesicarium isolates that cause Stemphylium leaf spot of spinach can originate from infected seed, as demonstrated previously for seedborne S. beticola (Hernandez-Perez 2005). If a seed crop is observed to have symptoms of Stemphylium leaf spot, it is advisable for the seed company that owns the proprietary crop to treat the harvested seed with an effective fungicide or hot water to reduce the amount of viable Stemphylium inoculum on seed.
There is a need to better understand the seed transmission risk of the pathogens that cause Stemphylium leaf spot. Hernandez-Perez (2005) demonstrated spinach seed transmission rates of 3.3 to 9.1% for Stemphylium spp. (3.7% to 10.3% of the seed on which Stemphylium spp. were observed) in greenhouse studies using misters to create ideal conditions for seed transmission, although that study did not account for the proportion of seedborne isolates that were not pathogenic to spinach (Hernandez-Perez 2005). Future studies should compare the incidence of the various Stemphylium spp. on spinach seed with the seed transmission rate under conducive conditions to determine the relationship between Stemphylium spp. incidence on seed and seed transmission risk. In addition, mark-release-recapture methods could be used, in which a spinach seed crop is inoculated with a tagged pathogenic isolate of S. vesicarium (i.e., an isolate with a unique MLG), the seed harvested and then planted as a baby leaf crop to observe for Stemphylium leaf spot, and isolates of the pathogen obtained from leaf lesions evaluated for the presence of the unique MLG. This could further elucidate the seed transmission risk of isolates of S. vesicarium.
References
du Toit, L. J, Hernandez-Perez, P. 2005. Efficacy of hot water and chlorine for eradication of Cladosporium variabile, Stemphylium botryosum, and Verticillium dahliae from spinach seed. Plant Dis. 89:1305-1312.
Hernandez-Perez, P. 2005. Seed transmission of Stemphylium botryosum and Cladosporium variabile in spinach. Pp. 79-101, in: Management of seedborne Stemphylium botryosum and Cladosporium variabile causing leaf spot of spinach seed crops in Western Washington. M.S. thesis, Washington State University, Pullman.
Hernandez-Perez, P., and du Toit, L. J. 2006. Seedborne Cladosporium variabile and Stemphylium botryosum in spinach. Plant Dis. 90:137-145.
Feng, C., Mansouri, S., Bluhm, B. H., du Toit, L. J., and Correll, J. C. 2014. Multiplex real-time PCR assays for detection of four seedborne spinach pathogens. J. Appl. Microbiol. 117:472-484.
International Seed Federation. 2017. Methods for the detection of Verticillium dahliae on spinach seed. http://worldseed.org/wp-content/uploads/2017/07/Spinach_Verticillium_July2017.pdf
Köhl, J., Groenenboom-de Haas, B., Kastelein, P., Rossi, V., and Waalwijk, C. 2009. Quantitative detection of pear-pathogenic Stemphylium vesicarium in orchards. Phytopathology 99:1377-1386.
Liu, B., and Correll, J. 2021. Discrimination of two closely related spinach leaf spot pathogens, caused by Stemphylium vesicarium and S. beticola based on species specific primers. Phytopathology 111:S2.30. (Abstr.)
Education and Outreach
Participation Summary:
Preliminary research results generated in this study were shared with western Washington and western Oregon spinach seed producers and the Pacific Northwest seed industry in a ~30-minute presentation at the Western Washington Seed Workshop (WWSW), held in conjunction with the Puget Sound Seed Growers’ Association (PSSGA) annual meeting and the Western Washington Small Seed Advisory Committee (WWSSAC) meeting on 13 January 2023. The presentation was titled "Stemphylium leaf spot of spinach: Research update". This workshop was attended by ~70 local seed producers, researchers, county extension leaders, graduate students, WSU technical staff, and representatives from all seed companies that contract with seed producers in western Washington. The SARE survey for this project was completed by attendees of the workshop.
On 22 February 2023, results from this study were shared at the Texas Spinach Field Day held near Crystal City, TX, and hosted by Tiro Tres Farms and Texas A&M AgriLife. In addition to a brief oral presentation, a 6-page handout summarizing results of Kayla Spawton's PhD research to date (2018-2022), including preliminary results from this study, was shared. The document was included in the full field day handout, and was titled "Stemphylium leaf spot of spinach: Susceptibility of cultivars to Stemphylium vesicarium, resistance of the pathogens to strobilurin (FRAC group 11) fungicides, and population genetics of the pathogen."
Results from this study were shared in a 15-minute presentation at the 10th International Spinach Conference on 1 May 2023 in Melbourne, Australia. The presentation provided an overview of Kayla Spawton's PhD dissertation, and is titled "Ecology and management of Stemphylium leaf spot of spinach". The international audience included ~120 spinach growers, seed company representatives, consultants, researchers, extension specialists, and students.
A poster was presented at the American Phytopathological Society Annual Meeting (Plant Health 2023) in August 2023 in Denver, CO. The poster focused on the results of this project, and was titled, "Genetic diversity in populations of Stemphylium vesicarium, causal agent of Stemphylium leaf spot of spinach, from leaves and seed." Attendees could view the poster over ~3 days, and seven attendees approached Kayla to discuss the study.
Results from this study were included in Kayla’s PhD Dissertation: Ecology and management of Stemphylium leaf spot of spinach. Results from Objectives 2 and 3 were included in Chapter 3: Association of pathogenicity with genetic diversity and population structure of Stemphylium vesicarium, causal agent of Stemphylium leaf spot of spinach (Spinacia oleracea). The results of Objective 1 were included in Chapter 1: Characterization of Stemphylium species associated Stemphylium leaf spot of spinach (Spinacia oleracea). In addition, the content of the dissertation was presented as an exit seminar to the Department of Plant Pathology at WSU, researchers from other universities, scientists at vegetable seed companies, and others (~50 attendees) on 18 Sep.
The Plant Health 2023 Conference was the only outreach activity in which the only topic presented was this specific project. The other presentations listed above included results from multiple aspects of the overall project on the ecology and management of Stemphylium leaf spot of spinach. This included presentations given at the International Spinach Conference in Australia in May 2023, and Kayla's exit seminar, in which the audiences primarily comprised scientists. In addition, two of the presentations were comprised primarily of producers: the 2023 Texas Spinach Field Day and the 2023 Western Washington Seed Workshop.
The survey was distributed to the audience at the WA Seed workshop. The majority of survey takers did not provide comments, but did select that the information was pertinent to them and will affect future decisions on managing the disease. However, it is unclear if that statement is based on the results from this specific project or based on other aspects of Kayla Spawton’s research project on the ecology and management of Stemphylium leaf spot, that were included in the presentation. Based on several comments from the surveys, the audience recognized how evidence of both pathogenic and non-pathogenic Stemphylium on spinach seed complicates the accuracy of current seed health testing, and acknowledged the importance of determining the pathogenicity of isolates before deciding whether a seed lot is highly infected with the causal agent of Stemphylium leaf spot of spinach. Constructive feedback included incorporating more real life examples that can make it easier for producers to understand how the results affect their businesses. Others stated the presentation was effective at sharing scientific information to a non-scientific audience.