Final report for LNC20-441
Project Information
As a perennial grain crop, intermediate wheatgrass (IWG, Thinopyrum intermedium, marketed under the trade name Kernza in reference to grain, and referred to as IWG subsequently) provides economic opportunity to growers who are interested in improving the environmental benefit of their farmland. Despite this opportunity, there is a possibility that perennial crops may harbor pathogen populations that could increase over time. Although IWG is not severely affected by many pathogens of annual crops, there are a few diseases that pose a threat to the long term viability of the crop and to human health. If the same pathogens also cause disease in widely grown annual crops, IWG might not be planted due to concern of spread. The fungus Fusarium graminearum, which causes the disease Fusarium head blight (FHB) is significant due to its production of the toxin deoxynivalenol, which is heavily regulated in food and animal feed. Viruses such as Wheat streak mosaic virus (WSMV) and Barley yellow dwarf virus (BYD) consistently appear across the Great Plains. WSMV, BYD, and FHB can cause tremendous economic loss to producers during epidemic years. We will work with farmers growing IWG to identify the pathogens present in their fields, measure the toxin content of the grain, use what we learn in their fields to inoculate and select plants resistant to infection in breeding nurseries on research stations, and test grain harvest, cleaning, and milling methods to reduce the toxin content of the grain. The outcomes of this project will be the expansion of a current grower network who will be able to identify and manage disease threats, development of disease resistant IWG populations, knowledge for growers about what diseases were found in their fields, and management recommendations for those diseases.
This project seeks to expand the IWG grower network to identify and address disease challenges in this new crop. In grower fields, we will identify Fusarium isolates, measure toxin levels in the grain, determine presence of viruses, and relate this information to growers with management recommendations. We would then use what we learn to establish a relevant disease screening program to develop improved seed for growers. The outcomes of this project include an expanded grower network, disease resistant IWG populations, information for growers about disease risk, and management recommendations for those diseases.
Perennial crops, including Intermediate wheatgrass (IWG, Thinopyrum intermedium) extend the growing season providing continuous living cover to stabilize eroding soil, reduce nutrient leaching and surface runoff, and sequester carbon. While policies and government programs encourage perennial plantings, these programs are rarely profitable. Kernza®, the trade name of IWG grain, offers growers a path to convert land and increase perennial acreage in a profitable way by producing a high value grain crop and additional biomass that can be grazed or hayed for feed.
IWG is immune or asymptomatic to many diseases of annual grains and has been used as a source of resistance genes for other cereal crops for barley yellow dwarf virus, leaf rust, stem rust, and stripe rust (Turner et al. 2013; Bajgain et al. 2019). However, IWG can be infected by Fusarium graminearum, a pathogen that infects the developing grain, causing Fusarium head blight (FHB) and producing a toxin compromising the value and safety of the grain. Serious FHB epidemics have occurred in 1982, 1990, 1993, 1995, 2008, 2009, 2015 and 2019 (Bockus et al., 2009; Dewolf et al., 2019; Rupp, unpublished) with annual losses averaging $6.5 million in wheat production. Approximately one million acres of wheat in eastern Kansas are annually at risk from FHB where rainfall is higher during heading and corn residue is more prevalent, providing high inoculum pressure and suitable environmental conditions for FHB development. Since 1980, wheat acreage in the eastern quarter of Kansas has declined by two thirds. A major cause of the decline has been aversion to the risk of FHB epidemics. Though the total number of acres devoted to Kernza (IWG) are small at this time, the value of the grain ranges from $3.69/lb to upwards of $6.50/lb for organic grain. Economic loss due to FHB for IWG growers could be great.
In research plots, IWG is largely resistant to screening isolates of Fusarium graminearum, but when planted in grower’s fields the level of toxin can exceed thresholds set by the FDA (Turner, unpublished data). In 2019, a KS statewide wheat Fusarium survey was undertaken by the Rupp Lab and collected isolates are being tested for their suitability to be used in future screens. It is a concern that the research isolates do not represent the races currently prevalent in Kansas or the Midwest, and may not be the same races that are infecting wheatgrass. Sampling IWG fields will allow identification of isolates and determination if they are the same as those infecting wheat. These isolates can then be used in breeding IWG and in making recommendations about crop rotation and disease management.
Wheat streak mosaic virus (WSMV; Family: Potyviridae; Genus: Tritimovirus) is one of the most common cereal viruses infecting wheat (Triticum aestivum L.) in the North American Great Plains (Burrows et al. 2015). WSMV has also been reported across the globe, causing sporadic epidemics on wheat (Navia et al. 2013). In the Great Plains, WSMV infections can cause wheat yield losses that range from 1 to 5% annually, however total loss in localized fields is common (Burrows et al. 2009; Ranabhat, N. personal observation 2019). Barley yellow dwarf (BYD) is another viral disease of concern that is ubiquitous in wheat production with variable losses (Ranabhat et al., 2020; Xu et al., 2020). As wheat acreage largely overlaps with IWG acreage, it is critical to assess risk to both crops.
Perennial crops living in fields for multiple years have a greater chance of exposure and retention of viruses than annual crops. Viruses show different symptoms depending on the crop or can appear asymptomatic, but still cause severe yield decline. While IWG has been used for conferring resistance to barley yellow dwarf virus (Bajgain et al. 2019) and identified as resistant to wheat streak mosaic virus (Cox et al. 2002), we do not currently know if IWG is infected with these or other viruses. Since controlling viruses in a perennial crop would be particularly challenging, it is important to proactively test for viruses that are widespread and devastating. One limitation of our methods is in focusing only on known viruses on related crops. While these economically significant viruses are important to the production of commercial crops like wheat, there may be unidentified viruses in IWG that we will not be able to capture that still pose yield risk to grain production. The use of Oxford Nanopore sequencing may overcome these challenges by allowing both pooling of large numbers of samples and full RNA sequencing that enables detection of unknown viruses (Fellers et al. 2019).
The idea for this project was sparked by processor and grower concern about the toxins produced by Fusarium and managing other diseases, which was expressed at the annual Kernza conferences held in Madison, WI in 2019 and Lindsborg, KS in 2018. Initial engagement in this project began through a grower network established in a Kernza biculture SARE project (2018) which is currently being expanded. The additional IWG farmers were identified by The Land Institute’s commercialization specialist as partners who might be interested in this particular project. The opportunity to build upon the already established grower network will facilitate increased grower/researcher connection and develop capacity to solve disease problems as they arise in a new crop.
Research
Our hypothesis is that Kernza is not immune to these diseases, but may instead be considered tolerant and a potential reservoir of pathogens.
After harvest in 2020 we will sample grain lots from each farmer’s field to test for deoxynivalenol (DON) toxin and to isolate Fusarium graminearum. Grain samples will be ground into a coarse flour and sent for testing at the North Dakota State University DON lab. The fungus will be isolated at Kansas State University in the Applied Wheat Pathology Lab (Rupp Lab). The Rupp Lab is an applied wheat disease laboratory that focuses on Fusarium head blight, among other fungal pathogens and viruses. Project co-coordinator Rupp leads the regional FHB mist-irrigated nurseries through the collaborative research network U.S. Wheat and Barley Scab Initiative. The isolates will be tested for ability to infect both IWG and wheat in the greenhouse over winter while determining aggressiveness and toxin producing ability. Macroconidia spore suspensions will be injected into heads that are near flowering. The inoculated heads will be covered with plastic bags for 24h following injection to ensure high humidity and aid in establishing successful infections. Disease severity will be assessed at 10, 15, and 20 days after inoculation. Susceptible and moderately resistant control wheat and IWG lines will be included in the experiment for relative disease severity comparisons.
Collection of F. graminearum field tissue will be performed according to protocols established by Leslie and Summerell (Leslie and Summerell, 2006) as follows. Symptomatic IWG heads will be collected, surface sterilized with 10% bleach, rinsed with sterile water, and plated on Spezieller Narhstroffarmer agar (SNA). Plates will be incubated at 20-25°C and 12h light/12h dark cycle. Following characteristic mycelial production, single macroconidia will be subcultured to produce a pure culture. DNA extractions from individual single-spore isolates will be used in PCR with established ITS (White, et al. 1990) and TEF-1 (O’Donnell, et al. 1998) primers to confirm species identity.
Isolates will be compared with those from nearby wheat fields using FHB survey data collected in 2019 and 2020 when available. Using information gained, we will generate inoculum using relevant isolates that can be used in breeding nurseries to select IWG plants that are resistant to toxin producing Fusarium races. FHB testing of breeding lines will be performed in misted field nurseries using soil-applied infested corn grain inoculum and in the greenhouse using single-floret inoculations. Visual disease evaluation methods will be used to rate the percentage spikelets infected by the pathogen and ground grain samples will be analyzed for the toxin DON. Viral symptoms will be evaluated during the growing season using visual phenotyping. Potential presence of unknown viruses may be explored through the use of Nanopore sequencing. Data will be disseminated to IWG producers and used by IWG breeders as they make selections for future varieties.These inoculated breeding nurseries, beginning in 2021 will adjoin already well-established FHB nurseries on research stations at two locations in Kansas. Inoculations which increase disease levels will not be conducted on farmer’s fields to avoid any economic hardship that the disease could have on the IWG or other crops.
Field inoculum consists of corn kernels infested with F. graminearum mycelium. Corn kernels will be soaked in water overnight, drained, and autoclaved to generate a sterile substrate. Seven to ten day old cultures of F. graminearum macroconidia in mung bean broth will be added to the sterile corn in breathable mushroom bags. The macroconidia will be allowed to germinate and infest the corn at room temperature on a 12h light/12h dark cycle. Mushroom bags will be agitated every 2-3 days for 8 days to break up clumping corn kernels and ensure even infestation. Infested kernels will be dried in the greenhouse and stored until needed at room temperature.
Approximately two weeks prior to heading, corn inoculum will be spread in the field. Misting irrigation will be used to provide adequate humidity and promote the production of perithecia on the kernels. The perithecia release ascospores into the canopy which will then infect the heads at the appropriate developmental stage (near anthesis, about Feeks 10.5), simulating a natural infection. Disease progression will be evaluated multiple times during head development and compared to growth stage notes taken during the season. Multiple wheat lines with known FHB reactions will be included within the experiment to serve as controls. Data will be analyzed using Dunnett’s multiple comparison test using a moderately resistant wheat line as the comparator for the reaction of an individual’s reaction to FHB.
Prior to the growing season in 2021, we will train growers and collaborators to be first detectors of the diseases of interest through Extension-style outreach. This includes identification of FHB, Wheat streak mosaic virus (WSMV), and Barley yellow dwarf virus (BYD). In the spring of 2021 and 2022, we would also collect leaf tissue and stem tissue, where viruses are most easily detected in wild perennial grasses (Lacroix et al. 2016), from IWG plants in farmer’s fields to test for multiple viruses that are common in other small grains, including the economically devastating WSMV, BYD, and others. This will be done through commonly used fungal culture, molecular (fungal and viral), and immunological (viral) techniques well established in the Rupp Lab. Collecting samples in season will give a good overview of the pathogens affecting IWG and serve as the first indicator how those pathogens may be interacting with annual crops in the production system. Growers will be provided instructions to send samples to the Rupp Lab and will receive results and potential management recommendations.
Samples of the grain collected from grower’s fields and experimental research stations will be analyzed for DON content. Healthy Food Ingredients will screen incoming IWG grain lots using ELISA. Samples with DON toxin levels above their limit will be sent to The Land Institute for research purposes. The grain will be dehulled at The Land Institute, in Salina, KS and milled at the University of Minnesota using a Brabender Quadrumat Junior Mill. The grain in the chaff, dehulled sample, bran, and flour will be tested for DON content to evaluate the amount of toxin that can be removed through dehulling the grain and milling the flour. At the research stations, individual heads will be harvested at three time points and the bulk grain will be harvested by both direct combining and swathing to determine if harvest timing and method affect DON levels in the harvested grain. Interested growers will provide samples from fields of different ages, intercrops, and management practices with potential to reduce the level of DON in the grain. The experiment station bulk samples and grower samples will be tested for DON toxin levels in addition to the experimental breeding lines.
Methodology Specific Citations:
Lacroix C, Renner K, Cole E, et al (2016) Methodological guidelines for accurate detection of viruses in wild plant species Running title: Virus detection in wild plants Christelle Lacroix. Appl Environ Microbiol 82:1966–1975. https://doi.org/10.1128/AEM.03538-15
Leslie, J.F. and Summerell, B.A. 2006. The Fusarium Laboratory Manual. Blackwell Publishing Ames, IA
O’Donnell, K., Kistler, H. C., Cigelnik, E., & Ploetz, R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences, 95(5), 2044-2049.
White, T. J., Bruns, T., Lee, S. J. W. T., & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications, 18(1), 315-322.
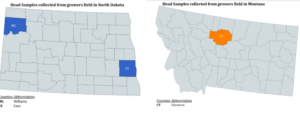
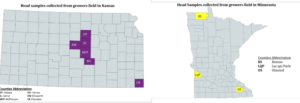
After harvest in 2020, 2021, and 2022 grain lots from each farmer’s field were collected to test for deoxynivalenol (DON) toxin and to isolate Fusarium graminearum (Figure 1).
Figure 1: Locations of origin of current samples
Grain samples were sent to The Land Institute in Salina, KS for DON and to Kansas State University's Applied Wheat Pathology (AWP) lab for fungal isolation. A graduate student was successfully recruited to work on this project for her M.S. She arrived in June 2021. Seventeen farmers and two researchers sent in grain samples. Additional samples were hand collected by project coordinators from both on-farm locations and disease nurseries. The AWP catalogued the grain samples, weighed approximately 10 grams, performed a visual assessment (%) of Fusarium Damaged Kernels (FDK), and saved seed for later virulence testing (Figure 2a and b).
Figure 2: Visual assessment (%) of Fusarium Damaged Kernels (FDK)
Due to the results found in 2021, ground samples were also analyzed for Fumonosin, a new addition. Symptomatic IWG heads were collected, surface sterilized with 10% bleach, rinsed with sterile water, and plated on Spezieller Narhstroffarmer agar (SNA). Plates were incubated at 20-25°C and 12h light/12h dark cycle. Following characteristic mycelial production, single macroconidia will be subcultured to produce a pure culture. DNA extractions from 69 individual single-spore isolates were used in PCR with established ITS (Figure 1 a.) (White, et al. 1990) and TEF-1 (Figure 1b.) (O’Donnell, et al. 1998) primers to confirm species identity in 2022. Results are detailed in Table 1 a-d. Samples that were also confirmed to produce toxin are labeled pathogenic.
Table 1. Fusarium confirmed samples from Kansas check with Fusarium specific primers
|
S.N. |
Sample ID |
Fusarium specific Primer check |
Fungal database |
Percent Identity |
Accession number |
Pathogenic |
|
1. |
19OTKS010 |
+ |
Fusarium incarnatum- equiseti species complex |
100%
|
GQ505636 |
|
|
2. |
18MCPKS011 |
+ |
Fusarium verticillioides |
98.05% |
MH582324 |
ü |
|
3. |
18HVKS012 |
+ |
Fusarium verticilioides |
99.34% |
MH582324 |
ü |
|
4. |
19SLKS013 |
+ |
Fusarium verticillioides |
99.10% |
MH582327 |
ü |
|
5. |
18CKKS014 |
+ |
Fusarium graminearum |
99.51% |
MH582244 |
ü |
|
6. |
20OTKS015 |
+ |
Fusarium verticillioides |
99.51% |
MH582324 |
ü |
|
7. |
19SLKS019 |
+ |
Fusarium graminearum |
99.24% |
MH582245 |
ü |
|
8. |
19SLKS020 |
+ |
Fusarium incarnatum- equiseti species complex |
99.66% |
GQ505612 |
|
|
9. |
19SLKS021 |
+ |
Fusarium boothii |
90.85% |
MH582232 |
|
|
10. |
19SLKS022 |
+ |
Fusarium boothii |
92.61% |
MH582231 |
|
|
11. |
19SLKS023 |
+ |
Fusarium graminearum |
98.65% |
MH582240 |
ü |
|
12. |
18CKKS025 |
+ |
Fusarium boothii |
93.71% |
MH582231 |
|
|
13. |
18CKKS026 |
+ |
Fusarium graminearum |
99.73% |
MH582244 |
ü |
|
14. |
18CKKS027 |
+ |
Fusarium armeniacum |
98.73% |
MH582290 |
|
|
15. |
18CKKS029 |
+ |
Fusarium graminearum |
100% |
MH582235 |
ü |
|
16. |
18CKKS030 |
+ |
No results |
|
|
|
|
17. |
18MCPKS031 |
+ |
Fusarium graminearum |
100% |
MH582235 |
ü |
|
18. |
18MCPKS032 |
+ |
Fusarium graminearum |
100% |
MH582235 |
ü |
|
19. |
18MCPKS033 |
+ |
Fusarium sp. |
100% |
GQ505596 |
|
|
20. |
18MCPKS034 |
+ |
Fusarium armeniacum |
100% |
MH582288 |
|
|
21. |
18MCPKS035 |
+ |
No results |
|
|
|
|
22. |
18MCPKS036 |
+ |
Fusarium armeniacum |
100% |
MH582288 |
|
|
23. |
17EWKS038 |
+ |
Fusarium acuminatum |
100% |
GQ505420 |
|
|
24. |
17EWKS039 |
+ |
Fusarium graminearum |
99.70% |
MH582244 |
ü |
|
25. |
17EWKS040 |
+ |
Fusarium armeniacum |
100% |
MH582288 |
|
|
26. |
17EWKS041 |
+ |
Fusarium incarnatum- equiseti species complex |
100% |
GQ505636 |
|
|
27. |
17EWKS042 |
+ |
Fusarium lacertarum |
99.41% |
GQ505593 |
|
|
28. |
17EWKS043 |
+ |
Fusarium incarnatum- equiseti species complex |
100% |
GQ505636 |
|
|
29. |
17EWKS045 |
+ |
Fusarium incarnatum- equiseti species complex |
100% |
GQ505636 |
|
|
30. |
21SLKS046 |
+ |
Fusarium incarnatum- equiseti species complex |
100% |
GQ505636 |
|
|
31. |
21SLKS048 |
+ |
Fusarium incarnatum- equiseti species complex |
100% |
GQ505636 |
|
|
32. |
21SLKS049 |
+ |
Fusarium incarnatum- equiseti species complex |
99.85% |
GQ505612 |
|
|
33. |
21SLKS051 |
+ |
No results |
|
|
|
|
34. |
20OTKS056 |
+ |
Fusarium incarnatum- equiseti species complex |
99.85% |
GQ505672 |
|
|
35. |
20MCPKS061 |
+ |
Fusarium incarnatum- equiseti species complex |
100% |
GQ505636 |
|
|
36. |
20MCPKS062 |
+ |
Fusarium armeniacum |
100% |
MH582288 |
|
|
37. |
20MCPKS063 |
+ |
Fusarium graminearum |
100% |
MH582235 |
ü |
|
38. |
20MCPKS065 |
+ |
Fusarium graminearum |
100% |
MH582244 |
ü |
|
39. |
21MCPKS067 |
+ |
Fusarium graminearum |
100% |
MH582235 |
ü |
|
40. |
21MCPKS068 |
+ |
Fusarium graminearum |
100% |
MH582235 |
ü |
|
41. |
21MCPKS069 |
+ |
Fusarium incarnatum- equiseti species complex |
100% |
GQ505636 |
|
|
42. |
21SLKS070 |
+ |
Fusarium graminearum |
100% |
MH582244 |
ü |
|
43. |
21MCPKS072 |
+ |
Fusarium incarnatum- equiseti species complex |
99.71% |
GQ505636 |
|
|
44. |
21OTKS075 |
+ |
Fusarium graminearum |
100% |
MH582244 |
ü |
|
45. |
20SLKS076 |
+ |
No Result |
|
|
|
Table 1 b. Fusarium confirmed samples from North Dakota check with Fusarium specific primers
|
S.N. |
Sample ID |
Fusarium specific Primer check |
Fungal database |
Percent Identity |
Accesion number |
Pathogenic |
|
1. |
20WLND016 |
+ |
Fusarium graminearum |
99.84% |
MH582248 |
ü |
|
2. |
21WLND066 |
+ |
Fusarium graminearum |
100% |
MH582247 |
ü |
Table 1 c. Fusarium confirmed samples from Minnesota check with Fusarium specific primers
|
S.N. |
Sample ID |
Fusarium specific Primer check |
Fungal database |
Percent Identity |
Accession number |
Pathogenic |
|
1. |
20OLMN017 |
+
|
Fusarium incarnatum- equiseti species complex |
100% |
GQ505636 |
|
|
2. |
19LQPMN053 |
+ |
Fusarium sp. |
100% |
MH582442 |
|
|
3. |
20LQPMN054 |
+ |
Fusarium incarnatum- equiseti species complex |
99.85% |
GQ505636 |
|
|
4. |
21LQPMN071 |
+ |
Fusarium graminearum |
100% |
MH582235 |
|
|
5. |
21LQPMN073 |
+ |
Fusarium incarnatum- equiseti species complex |
99.85% |
GQ505636 |
|
|
6. |
21LQPMN074 |
+ |
Fusarium sporotrichioides |
100% |
MH582277 |
|
Table 1 d. Fusarium confirmed samples from Montana check with Fusarium specific primers
|
S.N. |
Sample ID |
Fusarium specific Primer check |
Fungal database |
Percent Identity |
Accesion number |
Pathogenic |
|
1. |
21CHMT077 |
+ |
Fusarium acuminatum |
100% |
GQ505420 |
|
DON analysis revealed large differences between grower fields based on year and location (Table 2).
Table 2. DON testing results from 2022 by grower
|
Grower Cooperator |
DON(ppm) |
D3G(ppm) |
|
Don Wagoner Clearwater, MN |
0 |
0 |
|
B. Koffman- Gorrell |
0.1 |
0.1 |
|
B. Koffman- Lopez |
0.1 |
0 |
|
B. Koffman- Graber |
0 |
0 |
|
B. Koffman-Goering |
0.1 |
0 |
|
Clair Keene Williston, ND |
0 |
0.1 |
|
B. Koffman- Thompson |
0 |
0 |
|
Luke Peterson- Jt Farm |
0.9 |
0 |
|
Luke Peterson- Earthrise |
0.8 |
0.1 |
|
Estling Farm |
16.7 |
0.1 |
|
Chatfield, MN C5 Precleaning |
10.8 |
0.1 |
|
Chatfield, MN C5 post cleaning |
1.6 |
0.3 |
|
Chatfield, MN C5 post cleaning, grain |
1.5 |
0.3 |
|
B. Koffman- Gorrell-grain |
0 |
0 |
|
B. Koffman- Gorrell-hull |
0.4 |
|
|
B. Koffman- Graber-grain |
0 |
|
|
B. Koffman- Graber-hull |
0.1 |
|
|
B. Koffman-Goering-grain |
0 |
|
|
B. Koffman-Goering-hull |
0.1 |
|
|
B. Koffman- Lopez-grain |
0.1 |
|
|
B. Koffman- Lopez-hull |
0.4 |
|
|
Don Wagoner Clearwater, MN-grain |
0 |
|
|
Don Wagoner Clearwater, MN-hull |
0.1 |
|
|
B. Koffman- Thompson-grain |
0 |
|
|
B. Koffman- Thompson-hull |
0 |
|
|
Clair Keene Williston, ND-grain |
0 |
|
|
Clair Keene Williston, ND-hull |
0 |
|
|
Graber-Kernza Alfalfa-grain |
0 |
|
|
Bulk average Olathe |
14 |
|
|
Bulk average Salina |
0.86 |
|
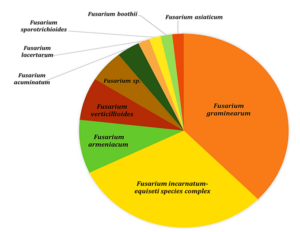
Figure 3. Diversity of Fusarium samples detected via PCR analysis using TEF primers
DNA sequencing and analysis is complete from these samples, however we are still collecting additional samples yearly.
Isolates will be compared with those from nearby wheat fields using FHB survey data collected in 2019 and 2020 when available. FHB testing of breeding lines associated with a misted field nursery located in Olathe, KS using soil-applied infested corn grain inoculum has occurred. The field site was expanded in 2022 to allow new entries. This nursery location is supported by SARE, but complementary nurseries through TLI are supported by FFAR to increase impact. Visual disease evaluation methods were used to rate the percentage of spikelets infected by a pathogen and ground grain samples will be analyzed for the toxin DON. Viral symptoms were evaluated during the growing season using visual phenotyping. In 2022, no viral symptoms were detected in the nursery. Inoculations which increase disease levels were not be conducted on any farmer’s fields to avoid any economic hardship that the disease could have on the IWG or other crops.
Prior to the growing seasons we continue to train growers and collaborators to be first detectors of the diseases of interest. Due to the Covid-19 pandemic, this has largely shifted to personal phone calls, emails, and Zoom meetings as most Extension-style gatherings have been severely limited, as well as sanctioned travel to neighboring states. This included the identification of FHB, Wheat streak mosaic virus (WSMV), and Barley yellow dwarf virus (BYD). In the spring of 2022, we collected leaf tissue and stem tissue, where viruses are most easily detected in wild perennial grasses (Lacroix et al. 2016), from IWG plants in farmer’s fields to test for multiple viruses that are common in other small grains, including the economically devastating WSMV, BYD, and others. Growers were provided instructions to send samples to the Rupp Lab and received results and potential management recommendations.
Samples of the grain collected from grower’s fields and experimental research stations will be analyzed for DON and due to the recent discovery of Fusarium verticillioides in this project, were tested for fumonisin content. Healthy Food Ingredients was unable to provide TLI with samples due to liability, but all collected samples will be tested by NDSU. Samples with DON toxin levels above the FDA’s will be dehulled at The Land Institute, in Salina, KS and milled at the University of Minnesota using a Brabender Quadrumat Junior Mill. The grain in the chaff, dehulled sample, bran, and flour will be tested for DON content to evaluate the amount of toxin that can be removed through dehulling the grain and milling the flour. At the research stations, individual heads will be harvested at three time points and the bulk grain will be harvested by both direct combining and swathing to determine if harvest timing and method affect DON levels in the harvested grain. Interested growers will provide samples from fields of different ages, intercrops, and management practices with potential to reduce the level of DON in the grain. The experiment station bulk samples and grower samples will be tested for DON toxin levels in addition to the experimental breeding lines. Grower submitted samples in 2020 and 2021, but the Olathe inoculated field was very high in DON content. However, in 2022, the inoculated sites were statistically more similar to grower fields.
Figure 4: Toxin testing results 2023
Nine different types of fungi, namely, Fusarium, Alternaria, Bipolaris, Rhizopus, Talaromyces, Epicoccum, Phlebiopsis,uncultured fungus and fungal sp. from the head samples were isolated. ITS1 and ITS4 primers were used as the sequencing primers for the genus differentiation in the beginning. Among the 79 sequenced samples, 55 were confirmed to be Fusarium species, 7 were Alternaria, 1 was Bipolaris, 3 were Rhizopus, 1 was Talaromyces, 4 were Epicoccum, 1 was Phlebiopsis, 5 were uncultured fungus and 2 were listed as fungal sp. when sequences were compared to NCBI database. Further confirmation of Fusarium sp. and Fungal sp. were carried out by amplifying the Tef-1α gene using Ef1 and Ef2 primers, which allowed us to confirm 10 different species of Fusarium among the 55 Fusarium confirmed samples and 2 fungal sp. samples. The sequences were compared to mycobank database ((https://fusarium.mycobank.org/). Unfortunately, one sample (18CKKS030) from the Fusarium confirmed samples did not generate a consensus sequence which might be because of the poor DNA quality and is not included in the list of Fusarium species confirmed. The most abundant species was Fusarium graminearum (n=21, 37.5%) followed by Fusarium incarnatum-equiseti species complex (n=17, 30.35%), Fusrium armeniacum (n=5, 8.92%), Fusarium verticillioides (n=4, 7.14%), Fusarium sp. (n=3, 5.35%), Fusarium acuminatum (n=2, 3.57%), Fusarium boothii (n=1, 1.78%), Fusarium lacertarum (n=1, 1.78%), Fusarium asiaticum (n=1, 1.78%) and Fusarium sporotrichioides (n=1, 1.78%) (Fig 2.6).
Among the 44 samples from Kansas, the dominant species isolated was Fusarium graminearum (n=15, 34.09%), followed by Fusarium incarnatum-equiseti species complex (n=14, 31.81%), Fusarium armeniacum(n=5, 11.36%), Fusarium verticillioides(n=4, 9.09%), Fusarium sp.(n=2, 4.54%), Fusarium asiaticum(n=1, 2.27%), Fusarium boothii(n=1, 2.27%), Fusarium acuminatum(n=1, 2.27%) and Fusarium lacertarum(n=1, 2.27%).
In addition, 6 samples were received from Minnesota and the most commom Fusarium species isolated was Fusarium incarnatum-equiseti species complex (n=3, 50%), Fusarium graminearum(n=1, 16.66%), Fusarium sporotrichioides(n=1, 16.66%) and Fusarium sp.(n=1, 16.66%) Likewise, 5 samples received from North Dakota contained Fusarium graminearum and 1 sample from Montana harbored Fusarium acuminatum.
The Land Institute of Salina conducted mycotoxin testing using ELISA(Agdia,) and lateral flow strip reader (Prognosis Biotech). The tests detected several mycotoxins, including DON (<0.2ppm), fumonisin(>3ppm), T-2/HT-2(<0.2ppm) and Zearalenone(<0.8ppm) which are known to be produced by Fusarium species isolated from the samples. The concentration of DON was lower than the rejection level for human consumption and there were detectable levels of fumonisin (>3ppm) and zearalenone identified as well (K. Turner, personal communication).
Kernza® is in the early phases of development and is a potential crop for sustainable agriculture. Research on this emerging crop is primarily focused on variety development and quantifying ecosystem services . Presence of ergot, Fusarium head blight, leaf blotch caused by Parastagonospora nodorum, and tar spot caused by Phyllachora graminis have already been reported based on the presence of fruiting bodies and DNA sequencing . These pathogens pose a threat for commercialization and adoption of the crop among producers. One of the main concerns is Fusarium head blight because Kernza® is a wild relative of wheat. Farmers face grain rejection from the commercial supply chain because Fusarium species produce mycotoxin in the grain. Some of these mycotoxins like DON, fumonisins, zearalenone, T-2/HT-2 toxins are regulated in food and feed. Since, there is limited information about diseases of Kernza®, the aim of this study was to examine fungal pathogens in the head and leaf samples, and to determine the diversity of Fusarium species present in the head samples in order to detect potential mycotoxins and ensure the safety of food and feed.
The result of this study found abundance of Fusarium species in the samples, while other fungi such as Alternaria, Rhizopus, Bipolaris, Epicoccum, Talaromyces, Phlebiopsis were also found in lower amounts. Fusarium was confirmed in 72% of the samples. Among Fusarium species identified, there were Fusarium graminearum, Fusarium incarnatum-equiseti species complex, Fusarium verticillioides, Fusarium armeniacum, Fusarium acuminatum, Fusarium lacertarum, Fusarium sporotrichioides, Fusarium asiaticum, Fusarium boothii and Fusarium sp. The causal agents for FHB are grouped within the Fusarium graminearum species complex FGSC).
Our findings identified 3 species from FGSC; Fusarium asiaticum, Fusarium boothii, Fusarium graminearum. Fusarium graminearum is the predominant pathogen to cause Fusarium head blight in cereals in the United States (X.-M. Xu et al., 2008). Fusarium incarnatum-equiseti species complex is reported to cause fusarium head blight in wheat, ear rot of foxtail millet and leaf diseases in leafy vegetables. Fusarium lacertarum is the member of Fusarium incarnatum-equiseti species complex which has been resolved by the multilocus sequencing technology of EF-1α gene, ITS+LSU 28s rRNA, RPB2 and CAM through genealogical concordance of phylogenetic species recognition (GCPSR) while there are other species in the complex which are still cryptic Fusarium lacertarum and Fusarium armeniacum are identified to cause FHB symptoms on susceptible wheat and sorghum heads. Fusarium verticillioides has been found to cause ear rot and blight in maize (Blacutt et al., 2018). Fusarium acuminatum causes root and crown diseases in wheat . Fusarium sporotrichioides causes ear rot in maize, root rot in soybean, and stem wilt in sea buckthorn.
Mycotoxin accumulation will be higher in severely diseased crop. Thus, it is a measure for aggressiveness of the pathogen. The mycotoxin testing done from TLI, Kansas in some representative samples of the study revealed that DON levels were below 0.5ppm, while fumonisin (>3ppm) and T-2/HT-2 toxin (>160ppb) concentrations exceeded recommended levels, and some of the samples also showed presence of zearalenone(>800ppb). It may not be economical to test for every mycotoxin but it is necessary to have knowledge of mycotoxins produced by isolated Fusarium species from the head samples and conduct tests for the regulated ones. This will be very useful in ensuring that the grains produced meet regulatory requirements, and protect farmers from grain rejection in the commercial supply chains in addition to ensuring food and feed safety. Growers and researchers will be more likely to implement disease management strategies and post-harvest practices to decrease the risk of mycotoxin contamination with accurate information about the potential for mycotoxin contamination in harvested grain.
Nanopore sequencing was conducted on high quality samples from the Olathe, KS nursery site. Sequencing results revealed the presence of Wheat streak mosaic virus, Barley yellow dwarf virus, and Soilborne wheat mosaic virus. Additional culturing of leaf spots revealed Tan spot, Spot blotch (Bipolaris sorokiniana + Tox A), and nodorum blotch.
The study was helpful to get the valuable information about head pathogens, and leaf pathogens of Kernza®. It provided an enhanced understanding of mycotoxins and their hazardous impact on both human and animal health. In the future, research could be focused on collecting samples from various states and utilizing DNA sequencing and mycotoxin testing to verify the presence of pathogens. Multi locus sequencing with various available primers can be done, along with a deeper examination of the equiseti-incarnatum species complex to distinguish them from one another. The identification of other pathogens such as bacteria and viruses, and their localization within the crop should also be investigated. Gene expression analysis in healthy and diseased crop can be done and it will be valuable for molecular work and breeding efforts. Root pathogens such as Rhizoctonia, Pythium are common problem in no-till systems (Cox et al., 2005). Given that root system, plays a critical role in the success of perennial agricultural system, understanding root pathogens is also essential.
Project Activities
Educational & Outreach Activities
Participation Summary:
Growers and researchers, as well as community members were able to attend field days. Additionally, farmers were able to attend an online webinar. A student was able to present her poster to the larger scientific community at Kansas State University. Upcoming presentations planned are two scientific meetings, one major regional grower meeting, increased social media presence, and an upcoming grower guide.
Learning Outcomes
- Disease detection, disease identification, sample collection
Project Outcomes
Fifteen growers reported actively scouting their crops for disease, and ultimately sending samples to be toxin tested.
Information Products
- Kernza Disease Guide Cards (Fact Sheet)