Progress report for LNE21-427R
Project Information
The long-term goal of our research program is to use natural genetic variation in tomato to increase sustainability of crop management systems by providing breeding tools for resistance to key pests and diseases of tomato. Septoria leaf spot (SLS) of tomato is becoming an increasingly important fungal disease in the Northeast and many neighboring states in the eastern seaboard of the US where hot and humid weather prevails during the growing season in most years. This aggressive foliar disease can devastate tomato crops, to which varieties with effective tolerance are not available. A recently released variety (‘Iron Lady’) was claimed to be tolerant to SLS, but our trials showed that it does not offer sufficient protection against the predominant pathogen strain in mid-Appalachia. There is a critical need of deconvoluting the genetics behind this trait and to develop resistant varieties via introgression of robust SLS resistance from wild species into commercial varieties of tomato. This research project, therefore, aims to respond to our farmers’ needs by identifying strong sources of resistance against SLS in the tomato germplasm, introgressing it into a cultivated variety, and creating breeding tools. The specific aims of this project are: 1) to introgress the novel trait ‘SLS resistance’ from wild accessions into a legacy tomato variety (e.g., ‘Cherokee’, 'WV-63') by creating interspecific hybrids via embryo rescue; 2) to identify the genetic locus responsible for SLS resistance; and 3) to develop bona fide CAPS markers for breeding. Backcrossing will be carried out in greenhouse conditions. Selection will occur in both, greenhouse as well as yearly in the field for performance evaluation at the WVU Organic Farm with the most advanced inbred line available. Previously, we circumvented a major bottlenecks of this project by identifying several sources of genetic tolerance (including complete resistance) to Septoria lycopersici in wild tomato accessions, producing viable hybrids, and we are introgressing the resistance into the cultivated tomato by recurrent backcrossing and selection. An advisory panel has been formed with crop specialists and farmers, who meet virtually with the research team for updates and discussion of the directions and priorities of the project. We have pre-built connections with farmers in WV and neighboring Appalachian regions, who will be actively involved in beta-testing and selecting the finalist genotypes prior to final selection for variety release. The main end-product generated from this research is a long-term, science-based sustainable solution for management of an increasingly important foliar disease to growers who will adopt this variety. The plant material and molecular markers developed in this project will be made available to farmers, breeders, and researchers.
As a direct response to farmers’ needs, the research objective is to develop a tomato cultivar that is resistant to Septoria Leaf Spot (SLS) by introgressing a novel natural resistance source we identified in wild tomatoes. No effective resistance against SLS is available in any tomato cultivar. We produced several F1 hybrids via embryo rescue. We will also map the resistance locus, identify the gene, and develop molecular markers for breeding. Our research will deliver a breeding toolkit to tomato breeders interested in incorporating SLS resistance in their cultivars, and the resistant materials will be available to farmers and breeders.
This project aims to address the threat the disease Septoria leaf spot (SLS) caused by the fungus Septoria lycopersici poses to tomato cultivation in the Eastern U.S., especially to organic farmers and home gardeners who cannot or are unwilling to spray fungicides on their tomato plants. A genetic solution by discovering genetic resistance to the disease and generating resistant plants that can be used by plant breeders is the most viable approach at the sustainability and economic standpoints. We have identified natural sources of genetic resistance to SLS in wild tomato accessions and generated hybrids through embryo rescue. This project aims at advancing the introgression work through recurrent backcrossing and selection for resistance until we obtain a useful variety with SLS resistance that can be directly used for tomato breeding. Moreover, we aim to develop molecular markers to facilitate breeding by identifying the locus (or loci) involved in this resistance.
Cooperators
- (Researcher)
Research
Research Goals: 1) Identify and introgress genetic resistance to Septoria leaf spot from wild tomato accessions into a cultivated variety, and make the genotype available to growers and seed companies; 2) Map the genetic resistance on the tomato genome and develop a molecular marker for selection (a bonus will be the identification of a culprit gene involved in the resistance mechanism).
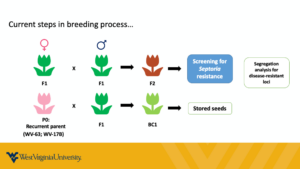
- Backcrossing of F1 hybrids with cultivated tomato (cultivar 'WV-63') to clean up the genome from non-domesticated alleles and introgress the resistance trait into Solanum lycopersicum. In later introgression stages, we will use the more conventional 'Cherokee' cultivar (or another variety chosen by the farmers, according to suggestions from the Project Advisory Committee) and will select not only for Septoria resistance but also for the other pathogens which the cv. 'WV-63' is uniquely known for being resistant to (Phytophthora infestans, Fusarium oxysporum, and Verticillium albo-atrum).
- Generation of segregating F2 populations to characterize the inheritance of the resistance (dominant/recessive, mono- or polygenic) and enable genetic mapping;
- Development of CAPS markers to map the resistance and, eventually, be used for marker-assisted selection (MAS) in breeding programs.
Year 1 report
 Backcrossing: BC1 has been completed for five F1 hybrids with cv. 'WV'63'. We are conducting a selection for Septoria resistance to identify the best materials (highest resistance levels) to move forward. Once the backcrossing/selection pipeline is well established and the graduate student is well trained since we conduct the backcrossing in the greenhouse, we expect to advance ~2.5 generations per year. We will switch to a more common variety (to be determined upon discussion with farmers in 2022) and select three more disease resistances from the BC3 generation on;
Backcrossing: BC1 has been completed for five F1 hybrids with cv. 'WV'63'. We are conducting a selection for Septoria resistance to identify the best materials (highest resistance levels) to move forward. Once the backcrossing/selection pipeline is well established and the graduate student is well trained since we conduct the backcrossing in the greenhouse, we expect to advance ~2.5 generations per year. We will switch to a more common variety (to be determined upon discussion with farmers in 2022) and select three more disease resistances from the BC3 generation on;
- Four F1 hybrids were evaluated in field conditions (WVU Organic Farm) in the summer of 2021. Last year was quite humid and hot, thus creating ideal conditions for the aggressive development of Septoria leaf spot. Indeed, the accessions of cultivated tomato cultivated in our organic plot (intentionally without any disease control) were devastated.
- Generation of an F2 segregating population: As expected, some F1 hybrids were self-incompatible and could not be selfed to generate F2 plants. however, we were able to produce pseudo-F2 populations by crossing different hybrids from the same genotypes (e.g., H1 and H3, both derived from crossings between cv. 'WV-63' and LA1984 (Solanum arcanum). We were also lucky to identify a self-fertile hybrid (H4: 'WV-17B' x LA1984). H4 was not only self-fertile but also the line that produced the most seeds and showed the highest resistance level. We have decided to move forward with segregation analyses with H4 for these reasons. Later, the other pseudo-F2 populations will be used to assess whether the genetic resistance behaves similarly when coming from other wild accessions. Characterization of genetic resistance in the H4-F2 population is currently underway.
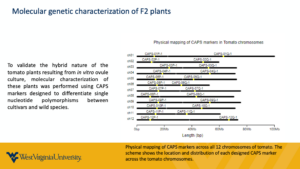
- Development of CAPS markers to map the genetic resistance to Septoria leaf spot: 24 markers were developed (one per chromosome arm of the tomato genome - this is possible due to the high degree of synteny in the genus Solanum). Markers have been designed to distinguish the domesticated chromosome arm and any wild tomato species under study (S. peruvianum, S. corneliomulleri, and S. arcanum) based on the genome sequence information available to us. The markers have been tested and optimized and are ready to be employed on the H4-F2 population to identify in which chromosome arm(s) the resistance is mapped. Later, we will develop markers to zoom in/refine the genetic locus with further development of markers on the identified arm to narrow down the locus further and further. We also used these CAPS markers to successfully confirm the hybrid nature of the lines we are using (and identified hybrid H5 as an escape, which only had the genome from the maternal progenitor, cv. 'WV-63').
Year 2 report:
-
In year 1 of the project, we focused on producing genetic resources to enable our research. We generated BC1 plants for advancing the backcrossing work to develop an SLS-resistance variety and developed CAPS markers for mapping the genetic resistance. We also evaluated the resistance level of each of the novel germplasm generated (cf. Y1 report above).
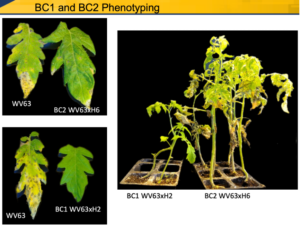 Expectedly, Year 2 was of intense work in the greenhouse with backcrosses and analyzing F2 progeny to map the genetic resistance. We faced challenges with the success rate of the backcrossing, but we were able to advance to BC2 with the H6 line, which generated plants with a good level of resistance. Plants showing the highest resistance levels (immunity to SLS) from 3 hybrids (H1, H2, and H3) are still in BC1 but advancing to BC2. One way we learned how to circumvent this genetic recalcitrance and speed up the backcrossing was to use the dwarf variety Micro-Tom as a bridge to advance to BC1. We have made strides on this approach and produced a population that is currently being screened for resistance and will be used to advance to BC2. We expect that at the end of Year 3, we will have reached BC4 with plants screened not only for SLS resistance but also for other fungal diseases (Phytophthora infestans, Fusarium oxysporum, and Verticillium albo-atrum) as well as developmental traits (plant size and architecture, flowering, fruit size, etc.)
Expectedly, Year 2 was of intense work in the greenhouse with backcrosses and analyzing F2 progeny to map the genetic resistance. We faced challenges with the success rate of the backcrossing, but we were able to advance to BC2 with the H6 line, which generated plants with a good level of resistance. Plants showing the highest resistance levels (immunity to SLS) from 3 hybrids (H1, H2, and H3) are still in BC1 but advancing to BC2. One way we learned how to circumvent this genetic recalcitrance and speed up the backcrossing was to use the dwarf variety Micro-Tom as a bridge to advance to BC1. We have made strides on this approach and produced a population that is currently being screened for resistance and will be used to advance to BC2. We expect that at the end of Year 3, we will have reached BC4 with plants screened not only for SLS resistance but also for other fungal diseases (Phytophthora infestans, Fusarium oxysporum, and Verticillium albo-atrum) as well as developmental traits (plant size and architecture, flowering, fruit size, etc.) -
A considerable amount of effort was placed into mapping the genetic resistance on the tomato genome. CAPS markers developed in the previous year were used to map a Ψ-F2 population generated from a crossing between H6 x H2 plants (due to self-incompatibility). The parental hybrids are both derived from S. peruvianum (accession LA2744). The analysis of 347 Ψ-F2 plants and 24 CAPS markers (2 per chromosome, one per arm) indicates that the resistance may be more complex than monogenic, especially because we observe segregation of the resistance level (perhaps a quantitative trait). We want to verify these results by conducting this analysis using a mapping-by-sequencing approach either on an F2 population from a different hybrid with the highest level of resistance observed (H4, which is the only self-compatible F1 hybrid from S. arcanum LA1984, and we now have enough F2 seeds available for the analysis) or on a more advanced backcrossing population (BC2 or BC3) derived from H4.

2022 Field Trial at the Organic Farm. Left: F1 hybrids; Right: tomato varieties showing susceptibility to Septoria leaf spot. The photo was taken in August 2022.
Given its hot and humid summer, 2022 was quite a challenging year in Morgantown, WV (and the whole East of the U.S.) for tomato plants cultivated without fungicide interventions. Our annual field trial at the WVU Organic Farm, which is cultivated without any spraying, demonstrated how devastating SLS is when left uncontrolled. Our disease resistance screening setup in the greenhouse is stringent as we utilize a more aggressive strain (from Long Island) than that occurring in Morgantown, WV, the inoculation level (50,000 spores/mL) and constant high humidity (almost 100% RH for more than a week). This setup aims to ensure that when a plant is scored highly resistant in the greenhouse, it will perform well in the field when the infection pressure is lower. In 2023, we plan to send BC1 and BC2 plants for performance evaluation in field conditions.
Year 3 Report:
As a recap, in Year 2, we conducted intensive greenhouse work, focusing on backcrossing and analyzing the F2 progeny for genetic resistance against Septoria leaf spot (SLS). Despite facing some challenges in achieving successful backcrosses, we progressed to BC2 with the H6 line, resulting in plants displaying robust resistance. The utilization of the dwarf variety Micro-Tom facilitated the backcrossing process as a bridge. Genetic resistance mapping of the tomato genome uncovered complexities potentially linked to quantitative traits.
In the past year (year 3), we overcame the challenges of crossings and advanced to the BC3 generation using the Hybrid-4 (which shows the highest Septoria-resistant level) in both 'WV-63' and 'Cherokee Purple' tomatoes as recurrent parents. Additionally, we advanced to the BC3 population with the remainder hybrids (H1, H2, H3, and H6). Our efforts for subsequent generations are now focusing on the H4, given its clear superiority regarding SLS resistance. In order to stack the resistance level as high as possible, we also conducted crossings between a genotype showing a high level of resistance in BC2derived from S. peruvianum (LA2744) (Ψ-F2 H6xH2) and a BC3 population from S. arcanum (LA1984) (H4) as a source of resistance. The plants from these crossings are being screened for Septoria resistance, and they will provide excellent insights into stacking or even increasing our current resistance level. We reached BC4 fruits using H4, which will be screened soon for SLS resistance, fungal diseases, and the developmental traits mentioned in the previous report.
The summer of 2023 in Morgantown, WV was unexpectedly dry, causing an effect of late disease symptoms in tomatoes cultivated following non-chemical practices. However, in late summer, when the conditions were more humid, we identified intense infestations of Septoria leaf spot and Verticillium wilt in our trials. This year, BC1 and BC2 plants were selected for performance evaluation under field conditions. Because of the stringent conditions of spore concentration, strain, and humidity under greenhouse conditions, the selected plants showed high resistance against SLS on the organic farm. Interestingly, a high level of resistance to Verticilium wilt was also observed in most of the plants, potentially pointing to a common strategy of genetic resistance. In 2024, we anticipate using BC4 and BC5 plants for field-based performance evaluation.
Our further exploration into mapping the genetic resistance in the Ψ-F2 population generated from a crossing between H6 x H2 plants, both derived from S. peruvianum (accession LA2744), included the QTL analysis using the Composite Internal Mapping (CIM) method. This approach allowed us to identify two markers among the initial 24 CAPS markers used (2 per chromosome). The position of the markers belongs to chromosomes 2 and 11. Additionally, 190 plants from backcrossing populations (BC1, BC2, and BC3) derived from H4 were also analyzed for QTL analysis via the CIM method. The results reveal strong evidence that the position responsible for the genetic resistance of SLS lies on chromosome 8, in addition to chromosome 11. Our efforts in mapping the regions responsible for SLS resistance allowed us to narrow down and identify a genomic region containing 34 genes within chromosome 8 after the design and evaluation of an additional set of 11 CAPS markers between the initial markers. The same strategy of narrowing down regions on chromosomes 2 and 11 is being applied with the design of new CAPS markers. Meanwhile, a fast advance in crossings is expected, and we expect to use a mapping-by-sequencing approach in a BC5 population derived from H4 to identify the limits of recombination and define a smaller set of genes potentially responsible for the resistance trait.
This is the report for year 3. We requested and have been granted a one-year no-cost extension of this project. The PhD student responsible for this project was recruited after about 6 months into year 1, and there was a learning curve, along with technical difficulties with genetic compatibility in advancing to BC1 and BC2. Now that all the technical issues have been resolved and the graduate student is sharp with plant care, phytopathology, and quantitative genetics, we anticipate that at the end of the next period we will have a tomato line with a strong level of resistance to Septoria (as well as other fungal diseases) and excellent growing and organoleptic properties. We also expect to provide markers and candidate gene lists within defined regions of the tomato genome potentially responsible for the genetic resistance to SLS.
Education & Outreach Activities and Participation Summary
Educational activities:
Participation Summary:
 The current pandemic certainly creates hurdles for in-person interactions and outreach activities. Nevertheless, we made efforts to have a presence with the farmers through participating in the WVU Organic Farm Field Day (a yearly event at WVU, which occurred on August 28, 2021 and August 27, 2022) by displaying a plot with our hybrids alongside other tomato cultivars. This year, the Septoria leaf spot disease was very aggressive on tomato cultivars (see picture 1), but impressively the five H1 hybrids we tested were mostly unharmed by them (see picture 2 - commercial varieties on the left and our F1 hybrids on the right). The plot was visited by around 30 local organic farmer families each year and counted with 10 agricultural educators and extension specialists.
The current pandemic certainly creates hurdles for in-person interactions and outreach activities. Nevertheless, we made efforts to have a presence with the farmers through participating in the WVU Organic Farm Field Day (a yearly event at WVU, which occurred on August 28, 2021 and August 27, 2022) by displaying a plot with our hybrids alongside other tomato cultivars. This year, the Septoria leaf spot disease was very aggressive on tomato cultivars (see picture 1), but impressively the five H1 hybrids we tested were mostly unharmed by them (see picture 2 - commercial varieties on the left and our F1 hybrids on the right). The plot was visited by around 30 local organic farmer families each year and counted with 10 agricultural educators and extension specialists.
In future years, after the pandemic is over, we aim to attend local and national in-person conferences, but for now, we will limit our participation in open-field and online to guarantee the safety of our researchers.
A Masters thesis by Estefania Tavares Flores (WVU Genetics and Developmental Biology Program, title: "Traditional and modern breeding strategies towards developing resilient crops: Two case studies in tomato"; advisor: Prof. Vagner Benedito) was defended in July 2021. Chapter II (Introgressing Septoria leaf spot resistance from wild tomato accessions into West Virginia cultivars using in vitro techniques and genetic markers: Developing alternatives for organic farmers) is a product of this research.
Year 3 Updates
We will present the results of this project at the 2024 Meeting of the National Association of Plant Breeders (NAPB) in Saint Louis, MO, and the Plant Health 2024 conference in Memphis, TN, both in July 2024.
Learning Outcomes
In our WVU Organic Farm Field Day, which occurred on Aug 28, 2021, and Aug 27, 2022 (each with some 30 farmer attendants) we were able to interact with 30 farmer families and 10 agricultural specialists, who were educated about Septoria leaf spot and other tomato diseases and could witness the strong disease resistance in our hybrid. Since this is year 1 of our project, we have not actively employed farmer participation but intend to do so for testing variety performance in later stages, when we have more advanced introgressions.
In 2022, we were also able to present our work to the Student Organic Seed Symposium (SOSS: https://organicseedsociety.org/meetings-events/student-organic-seed-symposium/) in July 29th (around 25 students attended the event).
We have also marked presence in the Food System Panel Discussion & Seed Swap: Sowing Sustainable Local Food Systems promoted by WVU (Eberly College, led by Dr. Mehmet Oztan).