Progress report for LNE22-457R
Project Information
We propose to develop and commercialize loop-mediated isothermal amplification (LAMP) assays for farm-based detection of six virulence marker genes from four Vibrio species associated with shellfish hatchery failures, along with an easy-to-understand Hatchery Instructions Guide for the assays. This proposal has been developed in response to an increased number of shellfish hatchery failures, which ultimately impact hundreds of oysters farms in the Northeast who depend on hatcheries for seed. Vibriosis, infection of larval or juvenile oysters by pathogenic Vibrio spp. is frequently a cause of these failures, reducing dependability of seed production. From a survey of hatchery managers, 86.7% indicated they would adopt the use of this assay into their hatchery operations to help diagnose production problems or verify that no pathogens are present in the algae they are growing as food for the larvae and seed. LAMP assays will be developed at Bigelow Laboratory for Ocean Sciences in Year 1 of the project, resulting in rapid, inexpensive molecular assays that can be implemented directly by hatchery operators, instead of sending samples out for expensive, long-turnaround PCR assays. The quick return of presence/absence of pathogens will let hatchery operators quickly develop management plans and mitigation strategies to remove the pathogens. The developed assays will be tested in the Mook Sea Farm (MSF) Hatchery in Maine, to verify that the protocols are appropriate for hatchery use. The MSF team will develop clear protocols and supplies lists in the Hatchery Instructions Guide, which will be shared with the Project Advisory Committee and a Hatchery Feedback Group to ensure the guide is user-friendly for those without molecular biology backgrounds. MSF and Bigelow Laboratory will seek commercialization opportunities for the custom LAMP assays, including but not limited to ‘kit production’, to ensure that high-quality LAMP assays are readily available to end users in a convenient, reliable, and contamination-free ‘easy to use’ format. Lessons learned through the development of these hatchery-focused LAMP assays will simplify the steps needed to develop LAMP assays for additional pathogens that are of concern to shellfish hatcheries and grow-out operations.
We propose to develop and commercialize loop-mediated isothermal amplification (LAMP) assays for farm-based detection of six virulence marker genes from four Vibrio species associated with shellfish hatchery failures, along with an easy-to-understand Hatchery Instructions Guide. The LAMP method allows for rapid, inexpensive, colorimetric detection of pathogens with minimal sample preparation, giving shellfish hatcheries straightforward diagnostic tools to manage hatchery production. These tools can be used to detect infections and determine management strategies, as well as to check shellfish seed viability prior to shipping to farms from the hatcheries, giving confidence to those purchasing the seed for their farms.
The oyster aquaculture industry depends on hatchery-produced seed. Northeast oyster farms annually purchase their seed from one of 40 Northeast hatcheries, which raise larvae and seed in land-based facilities, typically with 8-12 employees per hatchery. When a hatchery has production failure, farms lose access to seed that would have been their market product in 2-3 years, potentially putting thousands of jobs at risk. The seed supply for shellfish farming is increasingly challenged by hatchery failures (Walker 2017), with myriad potential causes (Jones 2006). Due to difficulty gathering production crash information from commercial hatcheries, recent research compiled production crash case studies from research hatcheries along the East Coast (Gray et al. 2022). Causes of crashes at these research hatcheries included vibriosis, infection of larval and juvenile shellfish by one of several pathogenic Vibrio species that cause mortality (Gray et al. 2022). When there is concern about pathogens, hatcheries currently must send samples to commercial laboratories to identify pathogenic Vibrios through PCR-based tests, waiting 2-4 weeks for results, at which point, the opportunity to control the pathogen has passed. These tests cost upwards of $200/sample, depending on how many genes are targeted, which can be cost-prohibitive for a small business losing product and sales.
This project will develop rapid, inexpensive loop-mediated isothermal amplication (LAMP) assays that can be performed onsite with basic equipment, allowing hatcheries to diagnose the presence of pathogenic Vibrio spp. within an hour, and immediately take proactive measures to control the pathogen spread. This project will develop LAMP assays to detect six virulence marker genes of four pathogenic Vibrio species known to cause production crashes at shellfish hatcheries (Gharaibeh et al. 2009, Ushijima et al. 2020, Xu et al. 2017, Yang et al. 2021). A recent survey to Northeast hatcheries elicited anonymous feedback such as ‘This work will certainly help shellfish hatcheries’ and ‘It will be a terrific asset for our industry.’ This technology represents a novel tool for shellfish hatchery pathogen management, and will likely be adopted across hatcheries to create more dependable production. That dependability will increase production efficiency, sustainability, and profitability for hatcheries adopting the approach, and a dependable seed source will increase efficiency and profitability for hundreds of oyster farms purchasing seed from these hatcheries.
Cooperators
- (Researcher)
- (Researcher)
- (Researcher)
Research
Pathogenicity varies between strains of identical species of Vibrio bacteria. Pathogenic strains of Vibrio are known to cause significant losses in oyster aquaculture. Timely and accurate detection of pathogenic strains is therefore necessary to detect and isolate infected seed and feed. This proposal seeks to address a need in the aquaculture community by developing a sensitive and rapid test for pathogenic Vibrio. We hypothesize that loop-mediated isothermal amplification (LAMP) methods will be ideal for detecting pathogenic Vibrio in aquaculture settings across three substrates: water, algae, and oyster seed.
In August 2022, our Project Advisory Committee (PAC) met to discuss the start of the project. The PAC consists of two hatchery operators, two farmer outreach specialists, and one technical advisor. We heard from the hatchery operators and one of the outreach specialist that in addition to the assays we are developing to detect hatchery pathogens, farmers would greatly benefit from LAMP assays to detect Aliiroseovarius Crassostrea (Juvenile Oyster Disease; JOD) as well as pathogenic strains ST36 and ST631 of Vibrio parahaemolyticus, which causes human health issues, as opposed to the oyster health issues caused by the Vibrio species targeted for our hatchery assays. As time and funding allows, we will aim to incorporate this into our work as well.
In September 2022, we used LampDesigner software to design LAMP assays for Aliiroseovarius (a target for which we already had samples, qPCR standards, and results to compare with LAMP) and Vibrio alginolyticus collagenase (clg) target. We ordered the primers and NEB WarmStart LAMP Fluorescent Master Mix and began testing on existing samples and a culture of Vibrio alginolyticus (B51) from the National Center for Marine Algae (NCMA) at Bigelow. Our LAMP reactions for Aliiroseovarius initially worked for the qPCR standard, but failed for the environmental samples. We returned to our alignment and noticed that the 3 prime end of the alignment, where LampDesigner had picked primers, was variable and could differ between the environmental samples and our standard. We then selected another set of primers in the more conserved 5 prime end of the region and tested these against our standard and environmental samples, with great success! For Vibrio alginolyticus we tested against the B51 culture and saw very strong amplification after just 10 minutes.
In October 2022, we designed endpoint PCR primers to isolate the collagenase (clg) target from the Vibrio alginolyticus B51 culture. This PCR product was then cloned into E. coli using the ThermoFisher TOPO-TA cloning kit. We then picked colonies, grew them to increase DNA yeild, and extracted plasmid DNA from the E. coli isolates. The plasmid was linearized by digesting with the NotI enzyme. This linearized plasmid was then serially diluted to be used as a positive control to measure the sensitivity of the assay.
In November 2022, we used the linearized plasmid of Vibrio alginolyticus B51 collagenase (clg) to determine the limit of detection for this assay, again using the NEB WarmStart LAMP Fluorescent Master Mix.
In December 2022, we used the NEB WarmStart LAMP Colorimetric Master Mix and a water-bath to test the Aliiroseovarius and Vibrio alginolyticus assays in conditions similar to a hatchery or aquaculture facility. We conducted these tests in triplicate to compare the sensitivity of the colorimetric assay with the fluorescent assay.
2023 Updates
Work on this project really accelerated in July 2023 with the addition of Ryan McCann to the team. Ryan is a student at the Roux Institute at Northeastern University pursuing a MS in Biotechnology and worked on this project from July through November to fulfill his co-op (internship) requirement for his MS. Using the workflow described in the 2022 report McCann was able to hit the ground running and began designing and testing LAMP assays for V. harveyi, V. coralliilyticus, V. splendidus, V. tubiashii, V. parahaemolyticus, V. vulnificus, as well as a general Vibrio LAMP that targets all species relevant to this project. Once those assays were designed, tested, and optimized, McCann focused his attention on an easy-to-use toolkit that aquaculture facilities could implement. We identified portable, self heating coffee mugs as a potentially cheap and accessible way to incubate LAMP reactions in hatchery settings. Further testing revealed that the "Piping" setting on the commercially available Nextmug maintains a steady 65 degrees Celsius, exactly the temperature needed for LAMP. The Nextmug can also aid in the DNA extraction step, which uses heat and mechanical bead beating to extract microbial DNA from hatchery samples.
McCann then tested the colormetric LAMP mastermix with the Nextmug, optimized a field based DNA extraction, and created a first draft guide for hatchery based LAMP experiments. In November 2023 We tested these materials at Mook's hatchery to see how they would perform and solicited feedback on improvements to the workflow.
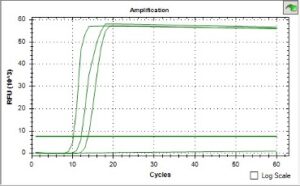
We have demonstrated the ability to design LAMP assays that are specific to the species of interest. These assays gave rapid results in as little as 10 minutes, demonstrating their potential for the aquaculture community. For Aliiroseovarius we used a dilution series of qPCR standards to determine that we are able to detect as few as 2,552 copies of the target, or 850 cells per reaction after 30 minutes, with further sensitivity testing ongoing. The sensitivity of Vibrio alginolyticus was similar to Aliiroseovarius indicating that the range of 1000-2500 copies per reaction is likely to be our limit of detection, though we hope to refine this as more assays are developed.
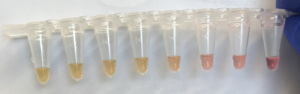
The colorimetric detection method that we tested gave a clear visual result in as little as 20-30 minutes using just a 65 C water-bath, and results were consistent with previous fluorescent detections. This indicates that these tests can be readily used in a hatchery setting. We plan to test small-scale, portable water-baths and conduct further hatchery testing in Spring 2023.
These early successes have energized our team and we look forward to tackling the next steps in the project.

2023 Updates
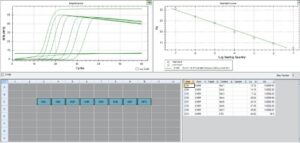
The seven assays that were designed in the fall of 2023 performed well when tested for specificity and sensitivity with positive controls. Several extraction methods were tested and all performed equally well so the team settled on the easiest and most cost effective method involving a single large ball bearing and an easy to make lysis buffer. These extractions, assays, and the first draft of the workflow were tested at Mook Sea Farm in December 2023 and January 2024 and showed promising results. The portable coffee cup gave very similar results to a thermal cycler, and results comparing fluorescent detection and colorimetric detection showed good congruence, though fluorescent detection may be better suited for samples with very low target concentrations; this is an area of research we plan to pursue in the early months of 2024.



Education & Outreach Activities and Participation Summary
Educational activities:
Participation Summary:
August 2022, Project Advisory Committee Meeting. We met virtually with our advisory committee and had a useful discussion around priorities for oyster hatcheries and growers.
November 2022, Peter Countway and Robin Sleith attended the Marine Genomics in the Gulf of Maine Symposium/Networking Event at the University of New Hampshire. This event brought researchers working on diverse genomic projects together to discuss problems relevant to the Gulf of Maine (and beyond). The subject of Vibrio pathogens came up several times and we were able to network with other regional partners studying pathogens.
In January 2024 Countway, Sleith, and Zimmerman attended the 2024 Northeast Aquaculture Conference & Exposition (NACE) and the 43rd Milford Aquaculture Seminar (MAS) in Providence, RI. Sleith presented the work that McCann accomplished in Fall 2023 and we gathered members of the advisory board for a brief check-in on our progress. We received good feedback on our project and have had follow-up conversations with the Bivalve Hatchery Health Consortium as we are planning to collaborate and include LAMP assays in the toolkits that are being distributed to hatcheries.