Final report for ONE21-390
Project Information
Vermont has a well-deserved reputation for innovation in sustainable agriculture, wonderfully illustrated in an award-winning Artisanal Cheese industry. This industry, similar to other niche food industries (e.g. wine, oysters), relies on the unique characteristics provided by the environments in which the raw materials are produced, a quality referred to as “Taste of Place” or terroir (or merroir for oysters). Much of the unique value of artisanal cheese is provided by the complex microbial communities present in raw milk, and how the processes of rind washing and aging shape these cheese microbial ecosystems. On the other hand, microbial communities in raw milk could also include pathogens affecting food safety, including Listeria monocytogenes, Salmonella spp. and Staphylococcus aureus. The goal of this partnership project was to improve protocols ensuring the safety, consistency, and organoleptic properties of artisanal cheese produced by The Cellars at Jasper Hill by evaluating the impact of feeding practices (hay versus pasture) on microbial communities in raw milk and cheese, as determined by a combination of culture-independent and -dependent methods. No differences in raw milk safety or quality profile or the microbial community of the cheese rind were seen between pasture or barn (hay) feeding samples, but these results should be interpreted with caution due to limitations in sample size. This research identified members of the microbial community in raw milk that were also present in cheese rind samples and enriched through the aging process. Metagenomics analysis of raw milk microbial communities identified infections by tick-borne parasites in the pasture herd, suggesting that this method could be used for health screening in the herd. Outcomes of this research include informed methods and protocols for evaluation of herd health and cheese and milk quality and safety at Jasper Hill Farms, and identification of candidates for microbes contributing to artisanal cheese flavor profiles.
The goal of this partnership project is to improve protocols ensuring the safety, consistency, and organoleptic properties of artisanal cheese produced by The Cellars at Jasper Hill. This project seeks to:
- Evaluate the impact of feeding practices (hay versus pasture) on microbial communities in raw milk and cheese, as determined by a combination of culture-independent and -dependent methods.
- Apply this information to investigate the potential relationship between microbial community composition and the safety and organoleptic properties of artisanal cheese.
- Foster knowledge exchange between partners, including: a) Advice on novel diagnostic methods for pathogens associated with raw milk; and b) Initial development of a comparative model (seafood – cheese) for the study of complex microbial communities in food.
The information developed in this research will aid Jasper Hill to develop a process for approving milk suppliers into their expanding artisanal cheese program.
Vermont has a well-deserved reputation for innovation in sustainable agriculture, wonderfully illustrated in an award-winning Artisanal Cheese industry (Paxson 2014). This industry, similar to other niche food industries (e.g. wine, oysters), relies on the unique characteristics provided by the environments in which the raw materials are produced, a quality referred to as “Taste of Place” or terroir (or merroir for oysters)(Teigen DeMaster et al. 2019). Much of the unique value of artisanal cheese is provided by the complex microbial communities (also called microbial ecosystems in this proposal) present in raw milk, and how the processes of rind washing and aging shape these cheese microbial ecosystems. On the other hand, microbial communities in raw milk could also include pathogens affecting food safety, including Listeria monocytogenes, Salmonella spp. and Staphylococcus aureus. The safety of artisanal cheeses to the human consumer is ensured by: 1) following farm practices that minimize or eliminate the presence of pathogen reservoirs in the farm system, and 2) screening the raw milk for pathogens of concern to humans. Therefore, the process of producing artisanal cheeses is both an art and a science, in which the raw ingredient (milk) is first screened for “microbial balance” (i.e. a diverse microbial community in which pathogens are minimized or absent), and then masterfully processed to produce safe and consistent cheeses with unique organoleptic profiles (Laithier & Percival, 2015).
There are many challenges to consistently achieving this delicate microbial balance in artisanal cheese. The microbial ecosystem in raw milk is influenced by numerous farming practices and environmental factors, including the feed, bedding, seasons, and climate, to name a few. There is, however, a limited understanding on how different farm practices contribute to the microbial communities in raw milk. By evaluating how specific farm practices at Jasper Hill and partner farms influence microbial communities in the raw milk used to make cheese, this proposed research addresses aspects of agriculture sustainability that are at the heart of the NE SARE mission: 1) “reduction of environmental and/or health risks in agriculture” through management of food safety in raw milk; and 2) “increasing farm productivity” by improving protocols at the farms that produce the milk and at the JH Microbiology Lab, ensuring the organoleptic quality and consistency of the cheeses produced by JH.
This proposed project was the product of conversations between Gomez-Chiarri and the JH team. Gomez-Chiarri, who, in addition to her passion for seafood, has a passion for artisanal cheese, was interested in deepening her knowledge in food microbiology and microbial ecology during a sabbatical leave. Inspired by a Jasper Hill event in which research on cheese microbiomes was featured, she approached Mateo Kehler to explore a potential collaboration. In this sabbatical, she hoped to expand her work studying microbial communities in marine organisms to the more controlled and repeatable microbial communities in artisanal cheese, while lending her expertise in genomics and development of diagnostic tools to the JH Microbiology Lab.
Citations:
Laithier, C., Percival, B., 2015. The Microbiology of Raw Milk: Towards a better understanding of the microbial ecosystems of milk and the factors that affect them., English. ed. Counseil National des Appellations d’Origine Laitieres and Specialist Cheesemakers Association, Great Britain.
Paxson, H., 2012. The Life of Cheese: Crafting Food and Value in America. Univ of California Press.Teigen De Master, K., LaChance, J., Bowen, S., MacNell, L., 2019. Terroir in Transition: Environmental Change in the Wisconsin Artisanal Cheese and New England Oyster Sectors. Sustainability 11, 2969. https://doi.org/10.3390/su11102969
Cooperators
- - Producer
- (Educator and Researcher)
- - Producer
Research


Objective 1: Evaluate the impact of feeding practices (hay versus pasture) on microbial communities in raw milk and cheese.
Jasper Hill purchased Andersonville Farm (Fig. 1, 2, with the goal of expanding their milk supply and determining if the production methods developed at Jasper Hill with a small herd of 45 Ayrshires can be scaled up to a larger operation. Andersonville Farm houses about 220 milking cows (Holsteins) that are fed on a Total Mixed Ration (TMR) diet consisting of dry hay, a grain mix, and whey from mid-September to mid-May. Part of this herd (about 2/3) was transitioned to pasture through Summer of 2021, returning to dry hay feeding in September.
In March 2022, we did a preliminary study to: a) practice the logistics of milk sampling at Andersonville, b) collect samples to optimize DNA isolation from milk; and c) compare the effect of dry cleaning cow teats with wood wool, a more ecological approach, as compared to iodine dips on microbial community (Figs. 3-5).



In early-May 2022, about half of the Andersonville herd continued on the current barn feeding regime, while the other half was transitioned to pasture. Herd feeding was performed by farm personnel under the supervision of the Farm Manager (Hunnewell) using the procedures currently used in the Andersonville farms. Confined cows were fed a Total Mixed Ration (TMR) including a blend of 1st and 2nd cut dry hay mixed with a mash of grain and minerals whetted with whey from Jasper Hill's cheese production. Cows on pasture were grazed on native mixed grasses and legumes, rotated on a 12-hour schedule. Pasture herds grazed for 12-hour intervals and were in the barn eating a TMR diet for the other 12 hours of the day.
Samples of raw milk from each of the two groups were collected 3 times (May 11, June 29, and September 9 2022, for a total of 12 samples - 2 groups x 2 replicate samples per group x 3 time points) for routine microbiological and milk quality testing and bacterial community composition analysis (Cosetta & Wolfe, 2019). Milk from each of the treatment group (dry hay TMR – named “barn” treatment hereafter, and “pasture”) was collected in a Double 8 rapid exit Delaval milking parlor (Figs. 4, 5). The milking parlor and equipment were cleaned between each of the two groups. Samples for microbiological analysis were collected from the milk bulk tanks using sterilized bottles for food safety and milk quality evaluation and transported to the microbiology lab at Jasper Hill for processing and storage or sent out to commercial laboratories.
Processing and Testing of Milk Samples
Samples of milk and filters from the milk lines were tested following routine culture procedures used in the Jasper Hill microbiology and associated laboratories, in order to detect and quantify the presence of targeted microbes known to affect milk and cheese safety and quality (e.g. E. coli, Staph, Listeria) using routine procedures (FDA’s Bacteriological Analytical Manual). These include plating of serial dilutions of samples on non-selective and selective media and bacterial identification using biochemical assays. A sample of milk from the bulk tank each group and milking time point was also frozen and sent to the University of Rhode Island for bacterial composition analysis).
Cheese Making
Milk collected at each time point (May, June, and September 2022) from each of the groups (barn and pasture) was used to make one batch of Alpha Tolman for each time and treatment group (Figs. 6 and 7). Alpha Tolman is an alpine type of artisanal cheese that ages for 8 – 9 months. Samples of cheese rind from a randomly selected cheese wheel from each of the cheese batches were collected at 4 times during the aging process using standard procedures (Cosetta & Wolfe, 2019) for a target of 144 total samples (pasture or barn treatments; May, June, September time points; duplicate rind samples from three wheels per treatment collected at 4 time points) for sequencing analysis of the microbial community. Cheeses were evaluated for organoleptic properties by Jasper Hill personnel, and ranked on a scale of 1 to 10 based on “deliciousness”. Notes on flavor profiles were also taken.


Analysis of bacterial community through metagenomic analysis
DNA isolation, library preparation, sequencing and initial bioinformatics analysis of the sequences was done by OneCodex (Wilmington, DE). Resulting data included taxonomic assignments using read counts, which were used to determine bacterial diversity (Shannon, Simpson indices) and composition (taxonomic relative abundance).
Objective 2: Investigate the potential relationship between microbial community composition and the safety and quality of artisanal cheese.
Sequencing data was transformed to account for the compositional data and analyzed using available statistical packages. Milk and cheese quality and safety were examined qualitatively (see limitations in results section below).
Overview of progress and description of changes to experimental approach
In the original proposal, sampling of milk from the Andersonville Farm was scheduled for summer 2021. Based on timing of the funding, the experimental design was revised in Fall 2021 to delay the start of the sampling to May of 2022. In January 2022, Gomez-Chiarri (URI) and Madelynn Johnston (grazing manager for Jasper Hill Cellars) visited the two farms directly involved in the project, Andersonville and Michaud, to discuss the logistics of the sampling process with the farmers. Information from the visits was discussed with the Jasper Hill Cellars team and was used to revise the sampling design (described in more detail in revised methods above). The team also brainstormed the type of environmental, milk nutritional and microbial profile, and cheese properties data to be collected at each sampling time point for each batch of milk and cheese. Based also on the visits and limitations in the resources at each farm, the team decided to focus the study on the Andersonville Farm.
Limitations of the study
There are several limitations to this study that should be considered when interpreting the data:
- Milking logistics at Andersonville Farm did not allow for replication in the collection of milk samples from the pasture and barn treatment groups at each milking time point.
- The cheeses were made as part of the usual production procedures. The following modifications had to be made to accommodate for issues experienced in the runs, leading to some limitations:
- One of two cheese batches (barn) made with milk collected in June had to be held back in aging due to differences in microbial profile, so the two batches (barn versus pasture) experienced a different aging schedule and environment.
- The milk samples from the September group collected for culture independent analysis were lost.
- The pasture cheese made from this milk batch was converted to another cheese type (Calderwood), based on a desirable flavor profile.
Therefore, we are not making any statistical inferences on the effect of treatment (barn versus pasture feeding) on raw milk and cheese quality. We focus the discussion of these results on the overall safety profile of artisanal raw milk and cheese made using the Jasper Hill Farms protocols and make recommendations on the potential use of metagenomics of raw milk bulk tank samples to evaluate (a) the food safety and quality profile of milk used to make raw cheeses and (b) the health of the herd.
Raw Milk Samples
Milk collection protocol and wood wool testing
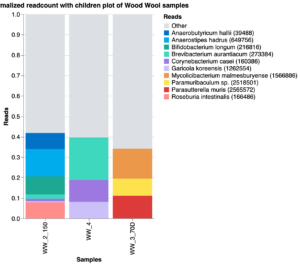
The milk collection protocol was tested in March 2022. The Jasper Hill team was interested in testing the use of wood wool, a dry and more environmentally sustainable method for cleaning cow teats before milking being used in the Swiss artisanal cheese. We determined, by treating five cows and milking them individually (as compared to bulk tank milk), that wood wool was less labor intensive than iodine dips, and that it had no significant impact on the microbiological food safety profile as determined using culture dependent methods (not shown). The metagenomic analysis only returned a low amount of reads for three of the five wood wool samples, and none for the tank control (Fig. 8), and no comparisons could be made between the iodine dip and the wood wool method. These very limited results from the culture-dependent assays are consistent with data from Europe indicating the potential for wood wool as an alternative to iodine dips, indicating that further studies may be warranted based on time savings.
Raw milk quality analysis
Pasture milk had significantly more protein (3.32%) than barn milk (3.16%; p=0.038), but less lactose (4.57% versus 4.61%; p=0.034). No significant differences (paired t-test) between the milk of pasture and barn cows were detected for percent butterfat, other solids, milk urea nitrogen, and fatty acid composition (not shown).
Presence of food pathogens in bulk tank milk
Culture-based microbiological analysis of the milk samples from each of the bulk tanks (n= 100 cows for barn and 110 for pasture group, with slight variation throughout the grazing season) did not show a decrease in the safety profile of the milk in the samples from cows fed the pasture regime as compared to the barn group (not shown). Milk samples from all these groups were determined to be safe for artisanal cheese making.
Indicators of mastitis from analysis of bulk tank milk
Somatic cell count in bulk tank, an indicator of mastitis infection in the herd, were not significantly higher (p=0.055; paired t-test) for the cows fed on the pasture regime (223,333 ± 20,817 cells/mL) than the barn regime (156,667 ± 25,166 cells/mL). These levels are similar to the milk-weighted geometric bulk-tank somatic cell count mean in the United States in 2019 of 171,000 cells/mL (USDA APHIS 2021).
The use of metagenomics to evaluate health of the herd
Metagenomic analysis of raw milk samples was dominated by host (i.e., cow) DNA, with only between 20 and 40% of the sequencing reads being of microbial origin (not shown). Interestingly, bulk milk from pasture cows collected in June showed the presence of pathogens that can cause disease in dairy cows, including the tick-transmitted pathogens Babesia ovata (33,347 reads out of about 2 million reads), Babesia bigemina (11,427 reads), and Theileria parva parva (4,128 reads). The presence of these tick-borne transmitted pathogens in a sample from the pasture herd and not in the barn herd is consistent with the increased risk of exposure to ticks in the pasture. However, this result indicating the presence of tick-borne pathogens only in pasture milk samples should be interpreted with caution, since this sample was also one of the two samples that returned the highest number of reads (around 2 million; the other one was one of the wood wool samples collected in March), while the number of reads from all other samples ranged between 149,734 and 874,430. A target depth in sequencing of at least 2 million reads is recommended for future use of metagenomics in the evaluation of raw milk food safety or herd health using milk samples. Moreover, a larger sample size of milk samples would be needed to make any conclusions on the relative impact of pasture feeding on milk safety. It is also impossible to determine how many cows were infected with these tick-borne pathogens in the bulk tank sample, but the small amount of reads detected in the bulk tank milk sample indicates that only a few of the animals were infected or that the levels of infection are low.
Cheese Rind Samples
Cheese Food Safety and Quality Profiles
Raw milk from cows from the Andersonville was used to make six Alpha Tolman cheeses at the Jasper Hill Cellars, one per each of the three milking time points (May 11, 2022; June 29, 2022; and September 7, 2022) and feeding regimes (pasture; barn). June samples were not included in the metagenomics analysis due to differences in storage conditions preventing direct comparison between the cheeses made with barn and pasture raw milk.
Expert testers from Jasper Hill Cellars rated the cheeses on a deliciousness factor, with 1 being the worst and 10 the best. Pasture cheeses were given a rating of 6 (May) and 7.5 (September), while the barn cheeses received a rating of 8 (May) and 7 (September). Each cheese had different tasting notes recorded, with no common notes shared for cheeses in each treatment group.
Bacterial composition of cheese rind samples
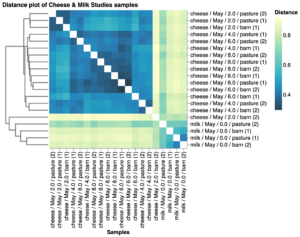
Beta diversity distance analysis of the microbial composition of milk and cheese rind for the May samples showed that samples cluster based on type of sample (milk versus cheese) and the length of time the cheese has aged (Fig. 9, the more distant the samples are in the plot, the more they differ they are in bacterial composition). These results confirm that the community of cheese wheels made from the same milk and aged in the same environment becomes more similar as cheeses age. No obvious clustering was observed between treatment (barn vs pasture) in this limited study.
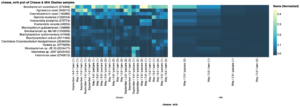
Several of these most abundant species in cheese rind were also present in the milk samples, potentially contributing to the unique flavor properties of artisanal cheese made with raw milk. The most abundant microbial species overall in cheese rind included Brevibacterium aurantiacum, Agrococcus casei LMG 22410, and Corynebacterium casei LMG S-19264 (Fig. 10). These three species, as well as others detected in this study, are commonly found in the rind of artisanal cheeses (Mounier et al., 2007; Wolfe et al., 2014).
Fungi are important members of the artisanal cheese community, contributing to the organoleptic properties of the cheese. The most abundant class of fungi (total amount of reads for all samples was 252,549) detected in cheese rind samples from Jasper Hill was Sacharomycetes (not identified to species), followed by Eurotiomycetes (several Aspergillaceae, including Penicillium biforme, P. roqueforti, and P. camemberti) (Wolfe et al., 2014). A very small amount of reads in cheese samples were classified to Protista (less than 10), Archeae (less than 700 overall), viruses (less than 2000), and mammalian host (less than 5000). None of the protistan detected in milk samples (e.g., the tick borne pathogens Babesia spp.) were detected in cheese rind samples.
Citations
Cosetta, C.M., Wolfe, B.E., 2019. Causes and consequences of biotic interactions within microbiomes. Current Opinion in Microbiology, Microbiota 50, 35–41. https://doi.org/10.1016/j.mib.2019.09.004
Mounier, J., Rea, M.C., O’Connor, P.M., Fitzgerald, G.F., Cogan, T.M., 2007. Growth Characteristics of Brevibacterium, Corynebacterium, Microbacterium, and Staphylococcus spp. Isolated from Surface-Ripened Cheese. Appl Environ Microbiol 73, 7732–7739. https://doi.org/10.1128/AEM.01260-07
USDA APHIS 2021. Determining U.S. Milk Quality Using Bulk-Tank Somatic Cell Counts, 2019. https://www.aphis.usda.gov/animal_health/nahms/dairy/downloads/dairy_monitoring/btscc_2019infosheet.pdf
Wolfe, B.E., Button, J.E., Santarelli, M., Dutton, R.J., 2014. Cheese Rind Communities Provide Tractable Systems for In Situ and In Vitro Studies of Microbial Diversity. Cell 158, 422–433. https://doi.org/10.1016/j.cell.2014.05.041
The pasture feeding regime did not increase the levels of pathogens in raw milk or the deliciousness of the cheeses made with the raw milk in this study. The quality of pasture milk was similar to that of barn milk, with the exception of a significant increase in percent protein and a decrease in percent lactose. This research showed how the milk contributes to the microbial communities of cheese rind made with raw milk, and how that community progresses through aging. Based on comparison with other research performed on microbial composition performed in European cheeses, Jasper Hill can use this data to identify some species that could contribute to unique flavor profiles in their cheeses, including several Penicillium spp.
Metagenomic analysis of bulk milk samples with a targeted read depth above 2 million could be used in monitoring for the health of the dairy herd and for the presence of potential food pathogens. Currently, there are several companies (e.g., One Codex) that offer easy, rapid, and competitively priced analysis of samples that include all steps in the process, from DNA extraction to data analysis and visualization. These analyses could complement current testing that is routinely done by Jasper Hill Cellars. The cost of these analyses is likely to go down in the future, and could be incorporated into the routine screening process.
Education & outreach activities and participation summary
Participation summary:
Results from the research were discussed at site visits and zoom meetings attended by the University of Rhode Island (Gomez-Chiarri, microbial ecology), Virginia Tech University (Petersson-Wolfe; mastitis) and members of the Jasper Hill Farms team (director, farm manager, grazing manager, microbiology and milk quality laboratory). Discussion topics included: (1) challenges with methods for the detection of pathogens associated with raw milk; (2) interpretation of the data from the microbial analysis; and (3) potential management recommendations. Some of these recommendations are being considered for implementation and inclusion in the standard procedures used by Jasper Hill to ensure herd health and product safety and quality. Further research will be needed to determine the impact of pasture and barn grazing on artisanal milk quality. The team is evaluating the presentation of results obtained in a peer-reviewed manuscript. Information and skill transfer also occurred from Jasper Hill Cellars to the University of Rhode Island Aquatic Pathology Laboratory, aiding in the use of a cheese rind model to study microbial interactions for seafood pathogens. Initial research was done on making a washed-rind cheese using oyster liquor, in which a marine probiont was used to shift the composition of the microbial community in cheese rind, decreasing the abundance of some genera and increasing microbial community diversity.
Learning Outcomes
- Informed methods and protocols for evaluation of herd health and cheese and milk quality and safety.
- Identification of some candidates for microbes contributing to artisanal cheese flavor profiles.
Project Outcomes
This project informed protocols for food safety evaluation of raw milk and the artisanal cheese made with this raw milk. Moreover, it pointed to a new method that could potentially be used to evaluate herd health. The project highlighted how the aging process selects for microbial communities that provide unique flavor properties to the cheese.
The experimental approach was too ambitious for the resources available at the farms (i.e., limitations in the ability to separate the herd during milking due to the limitation in the number of bulk tanks and milk trucks for milk collection). Additional technical challenges, besides limitations discussed in the results section, including isolation of DNA from microbes in milk. On the plus side, a decrease in the cost of sequencing allowed the use of metagenomics instead of 16S rRNA gene sequencing, allowing for identification of microbial at the species (and in some cases strain) level. Despite logistical limitations, data collected advanced knowledge on microbial communities in raw milk and cheese rind, complementing data obtained by Jasper Hill Cellars in collaboration with other researchers. Future research could explore the use of metagenomics in evaluation of herd health, and a more in depth examination of the microbial data obtained by metagenomics sequencing.
This research would not have been possible without the dedication of all the personnel at Jasper Hill Farms and Cellars, who contributed many hours and resources at no cost to the project. Mateo, thank you for sharing your amazing knowledge on artisan cheese making with the University of Rhode Island team and making this project possible! I also want to thank Christina Petersson-Wolfe for her generous contributions to the project, for sharing her knowledge in mastitis management with the team, and for critical evaluation of the experimental design and data. Many thanks for the Jasper Hill personnel for providing housing during our visits, to Julia Pringle for collecting and shipping cheese rind samples to the University of Rhode Island, Becky Klein for her tremendous insights into food safety, Nate Hunnewell for adapting milking procedures to accommodate the sampling, Maddie Johnston for compiling a wealth of data on grazing and milk quality, and the creamery and cellars teams for illustrating the art of cheese making and modifying their procedures to make special limited batches of cheese for this project.