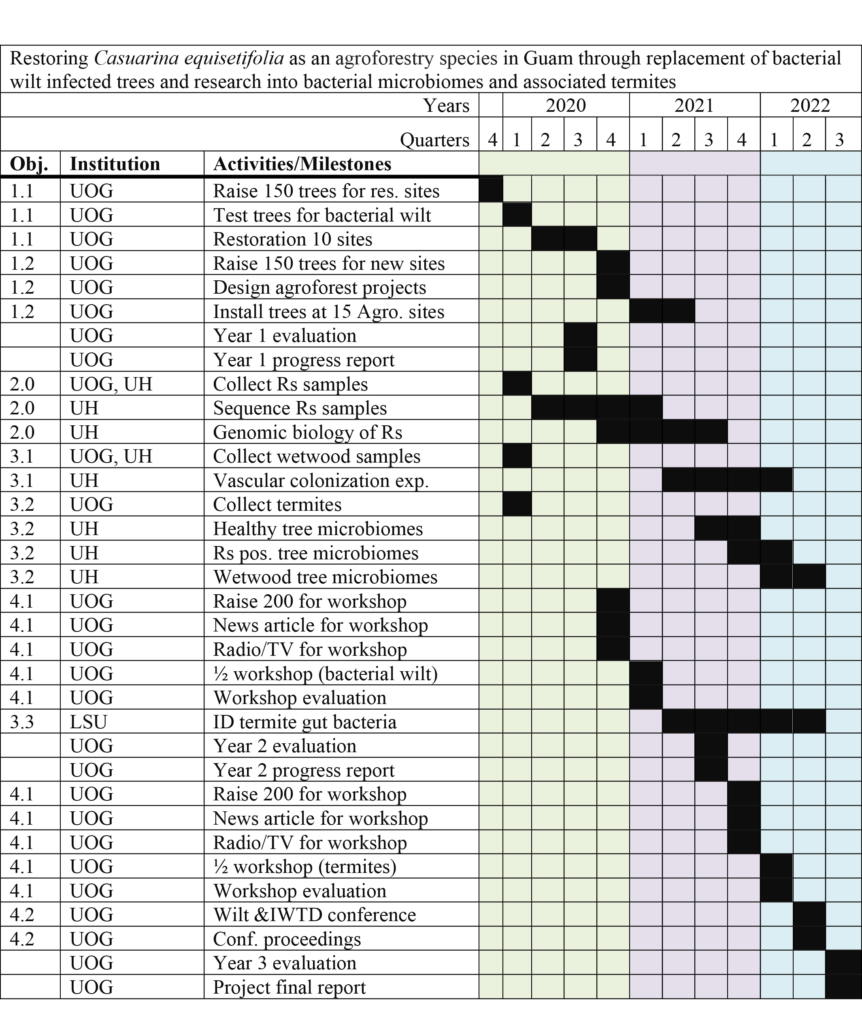
Final report for SW19-906
Project Information
Ironwood (Casuarina equisetifolia), recognized as an important tree species by the Natural Resources Conservation Service (NRCS), are in decline. In a survey funded by WSARE in 2008, it was estimated that 51% of the trees on Guam were showing signs of a progressive dieback now referred to as ironwood tree decline (IWTD). Between 2010-2019, data analyses identified 4 predictors of IWTD: the bacterial wilt pathogen Ralstonia solanacearum (RSSC) species complex, the butt and root rot fungus Ganoderma australe, termites, and bacterial wetwood. The importance of ironwood is underscored by the fact that it has been on Guam for thousands of years and is tightly integrated into the island’s environment and its many ecosystems. Ironwood is well suited to the shore lines of tropical islands, due to its ability to withstand salt spray, typhoon strength winds, and poor soil conditions. In Guam’s 2010-2015 Statewide Forest Resource Assessment and Resource Strategy report it was stated that ironwood decline was impacting the health of Guam’s forests and that determining its cause and finding solutions should be a priority for the future. Due to the similarities between IWTD in Guam and bacterial wilt caused by RSSC in China, the focus of this project was to examine the various biological components of RSSC and wetwood bacteria. Currently Guam is the only US location were RSSC has been confirmed in trees, whereas in China bacterial wilt disease occurs in Casuarina, Olea, Morus, and Eucalyptus. Some rank bacterial wilt disease as the most important disease in China due to its wide distribution and cumulative losses on trees, ornamentals, medicinal plants and many crops. The purpose of this project is to reduce the impact of bacterial wilt in Guam by (1) determining the origin of Guam’s ironwood tree bacterial wilt pathogen, (2) determining if the bacterium can be vectored by termites, (3) strengthen Guam’s ironwood tree population through increasing the genetic diversity of Guam’s ironwood tree population, and (4) educating the public and scientific community about bacterial wilt and IWTD.
Twenty-six water cultures of the bacterial wilt pathogen were collected on Guam and used in Sujan Paudel's 2020 thesis research at the University of Hawaii. In this study, he helped develop an efficient protocol for the isolation and characterization of RSSC strains from the declining trees. Both R. pseudosolanacearum (Rps) and R. solanacearum (Rs; strains were isolated from 2008 survey) were found to be associated with decline, although the latter species was found to be much less numerous (3) compared to the former species (35). The phenotypic characterization assays (Biolog) showed a similar utilization pattern for the Rps and Rs strains. The ironwood Rps population in Guam was found to be highly clonal, with the least nucleotide diversity and a contracting population structure. The MLST analysis identified North and Eastern Asia, Indonesia, and Northern Australia as potential origins of ironwood Phylotype I strains, whereas Central America, South-Eastern USA, Northern Latin America, and the Caribbean may be the potential origins of ironwood Phylotype II strains. The genomes of seven strains representing different phylogenetic groups were sequenced and annotated using long- and short-read sequencers. Ten more genomes representing different phylotypes were retrieved from the NCBI GenBank database. The reconstructed genealogy of the 17 Ralstonia strains using ClonalFrameML revealed a ratio of recombination and mutation rates equal to R/θ = 0.09592, an average length of recombined fragments δ = 22.3743, a divergence of DNA imported by recombination ν = 0.160891 and a relative effect of recombination and mutation r/m = (R/θ) x δ x ν = 0.345. The results indicated that homologous recombination had played a significant role in diversification/evolution of these strains. The most recombined strains were those from phylotype I within the lineage 3. The fastGEAR was used to identify ancient and recent recombination events across the core-genome of the 17 Ralstonia strains. Intriguingly, only one ancestral recombination event of 290-bp donated by the lineage 3 to lineage 2 seems to have occurred during the evolution time. Conversely, 572 recent recombination events were found across the population, which indicates that Ralstonia strains are still evolving; possibly to adapt to a constraining environment. A multiplex assay for specific detection and discrimination of reclassified Ralstonia species was also developed. The assay was used to identify Ralstonia from infected samples.
Termite samples collected on Guam for inclusion in Garima Setia's 2023 Louisiana State University thesis research were as follows: 42 Nasutitermes takasagoensis, 27 Coptotermes gestroi, and 6 Microcerotermes crassus. Garima’s thesis showed that none of the termite species (N. takasagonesis, C. gestroi and M. crassus) that attack ironwood trees in Guam were vectors for IWTD pathogens. The IWTD-associated pathogens were either absent or were scarcely detected in the termite worker samples collected from sick and healthy ironwood trees in Guam. Only Klebsiella sp. was detected in the worker samples of N. takasagonesis. Putative pathogens from genera Ralstonia, Klebsiella, Enterobacter, Pantoea, and Citrobacter were detected in low amounts (0.02% relative abundance) in the worker samples of C. gestroi. No pathogenic bacterial species associated with IWTD were detected in the worker samples of M. crassus. Bacterial communities of N. takasagonesis workers were found to be impacted by the presence of Ralstonia, tree health, plot average DS, plot average decline, proportion of dead trees in the plot, proportion of trees with termites in the plot, altitude, parent material, and site management, while those of C. gestroi workers were impacted by tree health and site management. The number of M. crassus samples was too limited to investigate the effects of those factors on the bacterial community. Feeding experiments were performed to investigate if wood consumption by termite workers was influenced by the bacterial load of ironwood. Ralstonia spp. were not detected in any of the N. takasagoensis samples, regardless if they were collected from trees with confirmed Ralstonia infection or healthy trees. One C. gestroi sample and none of the M. crassus samples showed Ralstonia. Four N. takasagoensis samples and one C. gestroi sample showed Klebsiella species, albeit in low amounts.
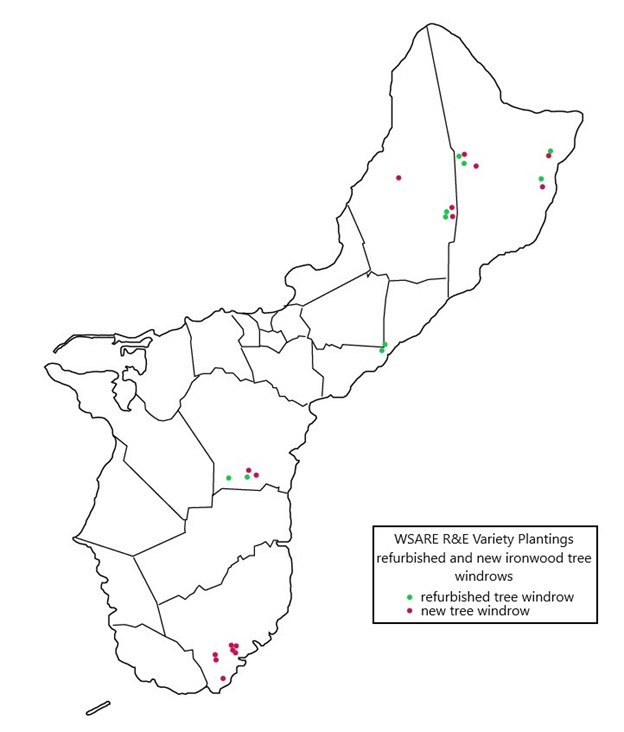
A total of 101 samples collected from 40 ironwood trees distributed across five geologically distinct locations on the island of Guam were sent to the University of Hawaii and used in Diksha Klair’s 2022 thesis research. Klair’s study focused on phenotypic metadata and 16S rRNA (V3) and ITS (ITS) amplicon-based microbiome analyses. The dominant bacterial and fungal phyla identified were Proteobacteria (75.6%) and Basidiomycota (61.44%), respectively. However, the average relative abundance of 24.04% of reads remained unclassified for fungal classification owing to a poorly annotated fungal database. At the genus level, Ralstonia was identified with higher richness from infected samples and, interestingly, greater abundance in shallow than in deep woody sample types. Irrespective of the other associated factors, the bacterial microbiota showed a close microbial association based on the relative abundance (disease severity) of Ralstonia, whereas the fungal microbiota formed an association based on identified Ganodermataceae. The study found significantly lower bacterial and fungal diversity and richness in Ralstonia-infected trees compared to healthy trees, putting plants' ability to host non-pathogenic endophytic microbiota at risk. In addition, conductivity, tree height, human impact, and the abundance of Ganodermataceae all had a significant impact on fungal diversity. Results suggested that deciphering the endophytic microbiota of ironwood trees and its association with Ralstonia will help to understand this complex pathosystem and can also act as a potential source for the formulation of biocontrol agents and the development of efficient disease management strategies. To increase the genetic diversity of Guam’s ironwood tree population, off-island varieties were planted in agroforestry projects. Fifteen new windrows consisting of 10 trees each were created, and ten deteriorating windrows were refurbished with the addition of 6-15 trees each.
To educate the public and scientific community about IWTD, three workshops were presented. In February 2020, two half-day workshops were attended by 36 participants, including farmers, property owners, home gardeners, professionals, and students at the University of Guam. In January 2022, an in-person and virtual ironwood tree decline (IWTD) conference was held for 3.5 days. In attendance were IWTD project researchers, IWTD research graduate students, and invited guests. Project researchers included Drs. Robert L. Schlub of the University of Guam, Claudia Husseneder of Louisiana State University, Zhong Chonglu of the Chinese Academy of Forestry, and from the University of Hawaii, Drs. Mohammad Arif and Shefali Dobhal. Graduate students reporting on IWTD research included Sujan Paudel of the University of Georgia, Garima Setia of Louisiana State University, and Dario Arizala of the University of Hawaii. The conference focused on the exchange of knowledge and research discoveries to ameliorate the impact of bacterial wilt in Casuarina equisetifolia, and unraveling the roles of Ralstonia solanacearum species complex, Ganoderma australe, wetwood bacteria, and termites in the decline of Guam’s ironwood.
In conclusion, this study provides research findings that are applicable for management of ironwood trees under the threat of bacterial wilt on Guam and other islands. The formation of ooze in cross-sections of large roots or branches is a strange indicator that a tree is infected with R. solanacearum or harmful levels of wetwood bacteria. Now knowing that high conductivity of water extracted drill shavings is a predictor of poor tree health, silviculturists have a quick, easy method to evaluate tree stress. This project provides evidence that Guam’s bacterial wilt pathogen most likely originated in China; therefore, trees that show resistance to this disease in China hold promise for Guam as well. From termite gut analyses, it was determined that termite workers are not vectors for the bacterial wilt pathogen or wetwood bacteria associated with IWTD; therefore, there is no evidence that controlling termite infestations will reduce the spread of Ralstonia. The findings suggest that Ralstonia infection reduces the plant’s ability to promote or possibly select beneficial endophytic microbiota. Linking population of beneficial endophytic microbiota with tree health will provide a means by which various tree management practices and environment conditions can be evaluated for their impact on overall tree health. Professionals and the general public are now being advised to reduce lawnmower and weed-trimmer damage to roots and the base of trees as a means to reduce infection and spread of pathogens. To reduce transmission of RSSC and wetwood bacteria when pruning, individuals are instructed to disinfect all tools. The public is also being advised to remove severely declined trees as a means to protect nearby healthy trees. We now know that most of the infected trees on Guam occur in cohorts of less than 12 trees but single infected trees are not uncommon. Even within tree cohorts, IWTD is slow to spread. It usually takes years from symptom onset to tree death. IWTD seems to be unique to Guam and has no single cause, though the majority of the tree loss can be contributed to R. solanacearum.
Obj 1 Yr 1-2: UOG Ed: Through tree plantings, educate the public on the importance of planting new off-island cultivars (seedlots from international provenance trails).
Obj. 1 Sub-obj. 1: UOG Ed: As part of the education effort, plant trees of mixed cultivars in 10 visually deteriorating windrows across Guam.
Obj. 1 Sub-obj. 2: UOG Ed: As part of the education effort, construct 15 new ironwood agroforestry projects using 150 trees consisting of a mixture cultivars.
Obj. 2 Yr 1-2: UH Res & UOG Res: Research into the bacterial wilt pathogen to determine the origin of Guam’s infection and its genomic biology.
Obj. 3 Yr 1-2-3: UH Res., LSU Res., & UOG Res: Research into the flora of ironwood trees and the guts of termites.
Obj. 3 Sub-obj. 1: UH Res. & UOG Res.: Research into the fungal flora of ironwood trees and their likely role in IWTD.
Obj.3 Sub-obj. 2: UH Res. & UOG Res.: Research into the bacterial flora of ironwood trees and their likely role in IWTD.
Obj.3 Sub-obj. 3: LSU Res. & UOG Res.: Research to determine if termites carry ironwood bacteria and thus, might be responsible for their movement.
Obj. 4 Yr 2-3: UOG Ed: Restoring ironwood as an agroforestry species in Guam through awareness and action of the local and scientific communities.
Obj. 4 Sub 1: UOG Ed: Through two ½ day workshops, attendees will learn about the care of the ironwood trees and its uses in agroforestry. Ironwood trees will be given away.
Obj. 4 Sub 2: UOG Ed, UH Ed & LSU Ed: PI and Extension/outreach representatives will conduct a four-day workshop/conference on bacterial wilt and other components of IWTD.
Cooperators
- (Researcher)
- (Educator)
- (Researcher)
- - Producer
- - Producer
Research
- The fungal flora of healthy and declining trees are significantly different.
- The bacterial flora of healthy and declining trees are significantly different.
- The bacterial flora of the guts of termites from healthy and declining trees are significantly different.
- There is a higher change of detecting Ralstonia in the gut of termites from trees that test positive for RSSC than those that do not.
- Ironwood RSSC isolates are from a single origin.
Obj. 2 Yr 1-2: UH Res & UOG Res: Research into the bacterial wilt pathogen to determine the origin of Guam’s infection and its genomic biology.
Methods: Obj. 2, University of Guam: research was conducted to find a method suitable for isolation of R. solanacearum from Guam’s ironwood trees. Though R. solanacearum could be detected from wood chips, drill shavings, water from root drill shavings, and stems and branches of trees, attempts to isolate from these same samples failed. The only means by which Rs could be isolated was by streaking ooze that formed on disks taken from stems, roots, or large branches of infected trees onto selective medium. To enhance the production of ooze, slices were placed on a saturated paper-towel in a moisture chamber for 24 hrs. Once formed, the ooze was streaked on Engelbrecht’s semi-selective medium (mSMSA) (Engelbrech 1994). Colonies were re-streaked onto SMSA, which was followed by streaking onto modified Kelman’s tetrazolium chloride medium (TZC) before grow-out on TZC (Norman and Alvarez 1989). Once grow-out plates had well-isolated fluidal growth, a transfer loop or spatula was used to transfer colonies to 3 ml of sterilized tap water in a 6 ml sterilized screw cap glass vials. The inoculation process is repeated as necessary to create a cloudy suspension. Duplicate vials were prepared, one for storage at room temperature in Guam and one which was shipped to the University of Hawaii for further evaluation.
Information recorded at each tree site included: tree tag number, date of collection, GPS location, visual decline rating (DS), site condition, height, DBH, altitude, geology characteristics, occurrence, root exposure, and presence of termites and Ganoderma wood rot. Tree drill shavings were collected from a single 3-inch deep hole using a ¼ inch drill bit. Shavings were assayed for R. solanacearum using R. solanacearum-specific immunostrip tests kits manufactured by Agdia Inc. of Elkhart, Indiana, USA (catalog number: STX 33900 and ISK 33900). Ralstonia solanacearum immunostrip testing was conducted on samples: either 1.5-3.0 mg of wood shavings or 80 µL of water from soaked drill shavings.
Methods: Obj. 2: University of Hawaii: water cultures from Guam were streaked multiple times onto TZC to ensure the cultures purity. A single colony from modified SMSA media was picked and mixed in 50 microliters of nuclease-free water. The colony was denatured for 10 minutes at 95oC and centrifuged for two minutes. The colony was used as a template to do endpoint PCR with Ralstonia solanacearum species complex specific primers (developed during this research; Paudel et al, 2022). DNA extraction was performed with cultures found to be positive with PCR. The DNA was extracted using a Qiagen kit. The identity was further confirmed by sequencing dnaA gene region. Sequencing was done with both forward and reverse primers. To infer the diversity, evolutionary relationships and genealogy of the ironwood decline strains, three housekeeping genes (dnaA, gap, gyrB) and two virulence related genes (hrpB and egl) were used.
Engelbrecht, MC (1994). Modification of a semi‐ selective medium for the isolation and quantification of Pseudomonas solanacearum. ACIAR Bacterial Wilt Newsletter Vol. 10: 3-5
Norman D, Alvarez AM (1989). A rapid method for presumptive identification of Xanthomonas campestris pv. diffenbachiae and other xanthomonads. Plant Dis. 73: 654-658
Paudel S, Dobhal S, Lowe-Power T, Schlub RL, Hu J, Caitilyn A, Alvarez AM, Arif M (2022). “RSSC-Lineage Multiplex PCR” assay detects and differentiates Ralstonia solanacearum, R. pseudosolanacearum, R. syzygii and the R3bv2 subgroup. BioRxiv. https://doi.org/10.21203/rs.3.rs-1693987/v1
A more in-depth description of the materials and methods for objective 2 can be found in the thesis of Sujan Paudel, UH graduate student: http://hdl.handle.net/10125/70339
Obj. 3 Yr 1-2-3: UH Res., LSU Res., & UOG Res: Research into the flora of ironwood trees and the guts of termites.
Obj. 3 Sub-obj. 1: UH Res. & UOG Res.: Research into the fungal flora of ironwood trees and their likely role in IWTD.
Obj.3 Sub-obj. 2: UH Res. & UOG Res.: Research into the bacterial flora of ironwood trees and their likely role in IWTD.
Methods: Obj. 3: Sub-obj. 1 & Sub-obj. 2: University of Guam: In 2021, ironwood trees from four locations across Guam were surveyed for suitability for the bacterial and fungal microbiome study. The trees and their sites were evaluated for several characteristics. A tree was classified as damaged if it had been cut into, drilled into, or otherwise damaged by humans (microbiome possibly or likely compromised); and undamaged if it had never been cut into, drilled into, or otherwise damaged by humans (microbiome uncompromised). Damage caused by natural phenomena (storm damage, water damage, or any damage not caused by direct human contact with the tree) was not considered. Trees were also examined for the presence or absence of termites, and presence or absence of Ralstonia solanacearum. Effort was made to choose an equal number of R. solanacearum positive and R. solanacearum negative trees for inclusion into the study. The size and health status of the trees were not specific as the main objective was to identify 30 ironwood trees which were damaged by human activity and 10 ironwood trees which were undamaged by human activity.
The following materials were used to execute the collection of woody ironwood tissue for the University of Hawaii
- GPS (Garmin or Cell Phone)
- Flagging Tape
- Tree tags
- Copper Nails
- Insight LH Precision Laser Rangefinder with Hypsometer
- Measuring Tape
- Tree Log Data Sheet
- Machete
- Hammer
- Wide chisel (or scraper)
- Nitrile gloves
- 1/4 in and 5/16 in drill bits
- Cordless drill
- Agdia Rs Immunostrip test strips and buffer filled bags
- Paper funnels
- Glass vials
- Ziploc bags
- Cooler
At each site, healthy and unhealthy trees were tested for Rs (R. solanacearum) using Agdia immunostrip tests. Drill shavings were collected from beneath the bark layer with a ¼ inch drill bit approximately 1.5 inches into the trunk of the tree. Approximately 0.2 grams of drill shavings were placed in Agdia test buffer extraction pouch. Testing continued until approximately five Rs+ and five Rs- trees were identified at each site. In some situations, Rs+ trees were unable to be identified at one site, so additional Rs+ trees were identified at remaining sites to make up the difference. If the tree had been uncompromised before testing for Rs, the test drill hole was filled with a sterilized stainless-steel screw, thereby conserving the integrity of the microbiome of the tree. After trees for inclusion in the study were identified, the following tree information was collected: tree tag number, tree GPS, location, tree disease severity, level of human impact, height, diameter at breast height, presence or absence of sporocarps, site condition, altitude, termite activity, and suspected termite species (Nasutitermes sp., Cototermes sp., or Microcerotermes sp.)
Conditions in the field were kept as sterile as possible: nitrile gloves were worn when preparing the tree and collecting the samples to avoid human skin cells in samples, machete and chisel used to clear off the bark were cleaned with Clorox wipes, all drill bits were autoclaved or flamed in the field, and all vials were autoclaved and wrapped sterilely to avoid contamination in transport. All samples were stored in the refrigerator (4°C) until shipment. Samples were shipped out the same day they were collected and overnighted to the University of Hawaii.
To collect woody ironwood tissue samples for the microbiome analysis, a protocol was developed to collected both shallow (0-3 inches) and deep samples (4-7 inches). At each depth, two combined samples were collected at 50 cm above the ground and on each side of the tree. Prior to drilling, a clean Clorox-wiped scraper was used to scrape off outer bark. When the bark was nearly removed, the scraper was cleaned with Clorox wipe again. Scraper was then used to slice off additional layers until the bark layer was easily separated from sapwood (Figure 1). If the exposed tissue appeared to be living, samples of drill shaving were collected, if not, another location on the tree was chosen.
Sampling began with the shallow woody tissue sample: drill shavings from a 3-in deep hole were collected using a 12 in by 1/4 in drill bit. At least 2 g (20 mL) of shavings were collected into a sterile glass vial by using a sterilized paper funnel (Figure 2). Vial with shavings was immediately placed on ice in the field for transport back to the lab. After the shallow woody tissue sample was collected, a 5/16 in by 12 in drill bit was used to enlarge the diameter of the hole and to increase its depth to 4 in. This would minimize the shavings contaminating the deep sample. For the deep sample, a sterile 12 in by 1/4 in bit was used to collect a total of 20 mL of drill shavings from the same drill holes, but at a depth of 4-7 inches into the tree. Again, shavings were collected into a sterile glass vial and vial with shavings was immediately placed on ice in the field for transport back to the lab. It should be noted that the depth of the deep sample was not allowed to exceed the radius of the tree. In these cases, depth was noted in the comments section.
In the same area of the tree (50 cm above ground) where the bark was removed for microbiome samples, additional holes were drilled and shavings collected for conductivity measurements. Conductivity is a measure of water's capability to pass electrical flow. This ability is directly related to the concentration of ions in the water. It is a significant predictor of percent wetwood within a tree. Greater conductivity is associated with greater % wetwood. Drill shavings were collected to the depth of 4 inches on both sides of the tree. Multiple holes were drilled with a 5/16 drill bit until approximately 75 mL of shavings were collected into a clean ziplock bag. The bag was placed on ice and transported to the lab. Once at the lab, samples were grinded until small enough to pass through a window screen (square opening of 3 mm). Five grams of sieved sample was then placed in a clean glass vial and placed in the freezer for storage 1 to 90 days. Ground shavings, which were powder-like and allowed to thaw out if necessary, were poured into 50 mL of distilled water in a 125 mL flask. The mixture was placed on a hot plate, brought to a boil, boiled for 5 minutes, then removed. While hot, mixture was then immediately run through filter paper. Filtrate was allowed to cool to room temperature before measuring conductivity. Values were recorded as microSiemens per centimeter (μS/cm at 25 °C). Conductivity level was then classified using the value: 0= slight (<301); 1= low (301-574); 2= moderate (575-849); 3= high (>849).
Six of the forty trees were chosen to be cut down (felled) for collection of additional microbiome samples: root sample, 150 cm felled sample, 300 cm felled sample, and 450 cm felled sample. Criteria included trees being undamaged (never having been drilled into, cut, or otherwise damaged by human activity), and half Rs positive/ half Rs negative. Two roots were sampled for each tree. When possible, each root had to be at least 2 inches in diameter. For each root, the spot where the root starts to go underground was marked (this might be directly at the trunk base or a foot or more away). Starting from the mark, the root was uncovered for a distance of 15 inches. Soil was removed from the exposed root using water and a soft scrub brush. At the 12 in distance, a drill sampling area was created by removing the bark with a disinfected machete. Using a 1/4 in drill bit, drill shavings were collected to a depth of 2 inches into each root by using a sterilized weigh boat to catch the shavings. The combined shavings were transferred into a glass vial until a volume of 20 ml was reached. Vial was immediately placed on ice in the field. Additional shavings were collected in a ziploc bag and tested for Rs using an Agdia immunostrip test.
After collecting the root sample, the trees were cut down (felled). The outer bark was removed from one side of the tree at distances of 150 cm, 300 cm, and 450 cm from the tree’s base using a disinfected machete (Figure 3, Figure 4). At each distance, drill shavings were collected to a depth of 2 inches using a sterilized weigh boat (Figure 4). The shavings were transferred into a glass vial until a volume of 20 ml was reached and then placed on ice to transport to the lab.
Figure 1. An exposed area of sapwood used for microbiome sampling.
Figure 2. Using a 12 in long by 1/4 in wide drill bit, drill shavings were collected into a sterile glass vial by using a sterilized paper funnel.
Figure 3. Felled tree showing 150 cm, 300 cm, and 450 cm collection sites cleaned and bark removed.
Figure 4. The 150 cm felled sample is collected using a sterilized weight boat (two people in front), while the 300 cm felled sample collection area has bark removed to sapwood in preparation for sample collection (one person in back).
Methods: Obj. 3, Sub-obj. 1 & Sub-obj. 2: University of Hawaii:
Genome sequencing of Ralstonia strains for evolutionary analyses: selecting ironwood strains from current and past Guam collections, their DNA was extracted from a half loopful of pure bacterial colonies, grown overnight on DPA (dextrose 5 g/L, peptone 10 g/L and agar 17 g/L) at 26 ± 2°C, by using the Genomic-tips 500/G kit (Qiagen) according to the manufacturer’s instruction. DNA concentration of the seven samples was measured using a Qubit 4 fluorometer (Thermo Fisher Scientific, Life Technologies, Carlsbad, CA); additionally, the DNA quality was assessed in a 1% agarose gel electrophoresis.
The isolated DNA were sent for illumine (NovaSeq) sequencing. Short-read sequencing of the seven Ralstonia strains was performed using Illumina NovaSeq system at the UC Davis Genome Center. DNA libraries were prepared using Seqwell plexWell LP384 Library Preparation Kit with 10 ng gDNA required for Illumina sequencing platform (seqWell, Beverly, MA). The prepared library was amplified with 8 PCR cycles, analyzed using Bioanalyzer 2100 (Agilent, Santa Clara, CA), and quantified by Qubit 4 Fluorometer Instrument (LifeTechnologies, Carlsbad, CA), and equimolar library pool was quantified and sequenced using qPCR with a Kapa Library- Quant kit (Kapa Biosystems/Roche, Basel Switzerland). Sequencing was run with paired-end 150 bp reads, using an and Illumina NovaSeq system. The sequences have been received.
The genomes were also sequenced using Oxford Nanopore MinION. DNA libraries were prepared using the Barcoding Kit (Oxford Nanopore Technologies Inc) with 400 ng of high-quality genomic DNA. The libraries were sequenced in a flow-cell version R10 and run for 72 hours. Sequencing was monitored in real time using the MinKNOWN software version 4.0.20 (Oxford Nanopore Technology). The generated FAST5 sequence files from Nanopore sequencing were base called using MinKNOWN software version 4.0.20 (Oxford Nanopore Technology). Hybrid assemblies were generated using both short- and long-reads, and evolutionary analyses were conducted. Core genome based evolutionary analyses was performed using ClonalFrame and fastGEAR software; total 17 genomes were used for analyses.
Bacterial flora Standardized DNA isolation method from ironwood plant: ironwood seeds sent by Dr. Robert Schlub in 2020 were sown for standardizing experiments and protocols. The critical and most important factor while sowing the ironwood seed was to place the seed horizontally 2-3 cm deep in the conical pots. The seeds germinate in 2-3 weeks depending upon the temperature condition. Ironwood seed germinated in 2 weeks under greenhouse conditions. The germination rate was lower comparatively to seeds sown in the month of January.
The modified protocol for DNA isolation from ironwood wood tissues was standardized.
DNA extraction from wood (Sapwood and heartwood) was quite difficult due to presence of higher quantity of secondary metabolites phenolic and lignin compounds. Therefore, after experimenting 8-10 different protocols we were able to finalize the protocol as follows:
Preparation of CTAB buffer (250ml):
- NaCl (2.5M) – Weigh 14.625 gm and dissolve in 100 ml of autoclaved distilled water
- Add Tris (1M) – 25 ml
- Add EDTA (0.5M) – 10 ml
- Add 5 gm of CTAB and stir until dissolved
- Make the volume up to 250 ml with autoclaved distilled water
- Adjust pH- 8.0 with conc. HCl
- Preparation of 10% PVP. Add 10 gm of PVP and dissolve in 100 ml of distilled water in a beaker. Filter using 0.22 μM filter membrane. Take a syringe of 10 ml, intake 8-9 ml of prepared solution at a time, fit the tip of syringe to 0.22 μM filter membrane without touching the surface of membrane, adjust the nozzle of filter to 50 ml centrifuge tube and gently press the syringe. Stored at room temperature.
Modified protocol for DNA isolation from greenhouse ironwood plants:
- Pre-heat 5 ml of CTAB buffer in water bath (65°C) in 50 ml centrifuge tube. Once it is heated, add 500 μl of 10% PVP and 10 μl of β-mercaptoethanol.
- Weigh 500 gm of plant tissue and grind in an autoclaved mortar pestle using Liquid Nitrogen. Grind well and make it in a powder form.
- Mix well the grinded tissue in the 5 ml of CTAB buffer prepared in step1.
- Use a wide borer tip (make a cut of around 1 cm on 1 ml tip using blade) to pour 500 μl solution from step 3 into bead beater tubes.
- Grind in the bead beater for 2 minutes at maximum speed.
- Incubate at 65°C for 1 hour and 30 minutes in a water bath. Vortex the tubes for 5 seconds after a 20-minute interval.
- Take out the tubes from the water bath and let it cool down for 2-3 minutes under.
- Add 5 μl of RNase (100mg/ml) and mix well. Incubate the tube at 37°C and 140 rpm for one hour in room 312 incubator with shaker.
- *Following steps to be done on DNA isolation workbench. Take aliquots of buffer solutions required for DNA isolation.
- *Buffers used in the following steps are taken from Qiagen DNeasy Plant Mini Kit (Catalog no. 69104)
- Add 130 μl of AP1, mix well with pipette and incubate in ice for 5 minutes.
- Centrifuge the tubes at 20 X1000 g for 5 minutes.
- Pipet the lysate into a QIAshredder spin column placed in a 2 ml collection tube.
- Centrifuge for 2 min at 20 X1000 g.
- Transfer the flow-through into a new tube without disturbing the pellet if present. Add 1.5 volumes of Buffer AW1 and mix by pipetting.
- Transfer 650 μl of the mixture into a DNeasy Mini spin column placed in a 2 ml collection tube. Centrifuge for 1 min at ≥6000 x g (≥8000 rpm). Discard the flowthrough. Repeat this step with the remaining sample
- Place the spin column into a new 2 ml collection tube. Add 500 μl Buffer AW2, and centrifuge for 1 min at ≥6000 x g. Discard the flow-through.
- Add another 500 μl Buffer AW2. Centrifuge for 2 min at 20,000 x g. Note: Remove the spin column from the collection tube carefully so that the column does not come into contact with the flow-through.
- Transfer the spin column to a new 1.5 ml or 2 ml microcentrifuge tube.
- Add 25 μl Buffer AE for elution. Incubate for 5 min at room temperature (15–25°C).
- Centrifuge for 1 min at 8000 g.
- Repeat step 17
- Store DNA in aliquots at -20°C.
The current study focused on the collection of ironwood samples associated with ironwood decline, along with phenotypic metadata, and 16S rRNA (V3) and ITS (ITS1 and ITS2) amplicon-based microbiome analyses.
This research involved a large-scale sampling (101) of ironwood tree samples from 5 different regions on Guam, a geographically isolated and unexplored island with unique climatic conditions, with the aim of resolving ironwood decline-Ralstonia disease complexity and revealing endophytic microbial interactions within the host system. The comparison of endophytic microbial community associated with different factors such as Ralstonia infection, root rot fungus, disease severity, termite activity, etc. considered in this study, will not only pave a way to resolve disease complexity but also will provide an insight into the complex interactions occurring among endophytic microorganisms sharing the same niche and their role in modulating pathogen response. Three characteristics were purposefully replicated, one being the detection of Ralstonia using immunostrips. Roughly half of the trees tested positive for Ralstonia and half tested negative. The other two characteristics replicated were location and tree disease severity.
The samples from Guam were processed immediately for DNA isolation using the modified method. The first run was with an in house 16S primer set to check the quality. The DNA isolated from 101 samples was sent to the Microbial Genomics and Analytical Laboratory (MGAL) core facility at the University of Hawai'i at Mānoa for 16S and ITS library preparation. Community analysis was performed using EzBioCloud. The Kruskal-Wallis H test (α=0.05) was used for statistical comparison of alpha diversity identified by bacterial and fungal richness (total number of genus) and diversity index (Simpson’s index) associated with A) Ralstonia infection and B) sample type.
A more in-depth description of the materials and methods for this objective can be found in the Diksha Klair, University of Hawaii Master of Science thesis: https://hdl.handle.net/10125/103907
Obj. 3: UH Res., LSU Res., & UOG Res: Research into the flora of ironwood trees and the guts of termites.
Obj.3 Sub-obj. 3: LSU Res. & UOG Res.: Research to determine if termites carry ironwood bacteria and thus, might be responsible for their movement.
Methods: Obj. 3, Sub-obj. 3, University of Guam:
Termite Collection Methods: from 2019-2021 termites were collected on the island of Guam. For each ironwood tree with an active infestation, general data was recorded, including: Tree tag number, date of collection, GPS location, visual rating, site condition, height, DBH, altitude, geology characteristics, occurrence, root exposure, collection area of termite ( i.e. mud trail, above or below ground colonies), and presence or absence of Ralstonia solanacearum (tested using Agdia Immunostrip buffer test pouch). A minimum of 21 active termites (15 workers/6 soldiers) per tree were collected directly from the infestation site with a general aspirator and immediately transferred into vials. For each tree, the samples including soldier and worker termites which were partitioned into 10 mL of 70% ethanol (for morphological identification) and 10 mL of 95% ethanol (for Illumina sequencing). Each individual vial was labeled by tree number in the field, placed in a vial box, then placed on ice in a cooler. The vials were further processed through labeling by clinic number, then in a cardboard vial box with the lid closed and placed in the laboratory freezer.
Termite-sampled ironwood trees were tested for Ralstonia solanacearum by selecting two locations on the stem breast height above the soil line. An electric drill with a 1.5 in. sterilized drill bit was used to create the initial break in the periderm, then a 5/32 in. sterilized drill bit was used to collect drill shavings by drilling a 4 cm deep hole in the tree. The shavings were collected in plastic bags and subsequently labeled. To reduce the chances of secondary infection by other insects or pathogens, the two holes were plugged with a sterilized Flat Head #12 (7.32) stainless steel screw. The shavings were then brought back to the University of Guam Cooperative Extension Plant Pathology Lab and tested for Rs using the Agdia® Inc. Ralstonia solanacearum (Rs) immunodiagnostic strips. The test was performed on the same day the samples were collected. Following the manufacturer’s instructions, 0.15 g of the drill shavings were placed in BEB1 sample extraction bags. The sample was allowed to set three minutes before inserting the immunostrip.
Drill shavings were collected for conductivity determinations. In the case of LSU, the bark was removed just to the point that the live tissue layer appeared (phloem) before being drilled and shavings collected. In the lab, tissue samples were grinded and sieved through 1/8 in galvanized hardware cloth. Five grams of sieved tissue was then placed in a clean glass vial and placed in the freezer for storage. Processing method included the following: the entire 5 g of sieved tissue was transferred into a 125 mL flask containing 50 mL of steam-distilled water, shaken, and placed on a hot plate. At first sign of boiling (approximately 2 minutes later), mixture was boil rapidly for an additional 5 minutes. While hot, using a Vortex-genie 2, the sample was vortexed vigorously for one minute at the medium setting. Afterwards, it was placed in an ice bath and cooled to 60°C, then filtered through Whatman #3 paper by pulling a vacuum. After the filtrate cooled to room temperature, its conductivity was measured using a Hach 51800-10 sensION 5 Waterproof Conductivity Meter and recorded as microSiemens per centimeter (μS/cm at 25 °C). Conductivity level was then classified using the value: 0= slight (<301); 1= low (301-574); 2= moderate (575-849); 3= high (>849).
Soil Collection Methods: Using the same ironwood trees that termites had been collected from, in 2022 five core soil samples were collected per tree and combined for processing. Using a clean, disinfected handheld soil core sampler with an inner diameter of ¾ inch, five soil cores were collected into a labeled zip lock bag. The collection of five samples was roughly equally spaced around the tree and 3-5 ft from the tree base. Without disturbing the underlying soil, any grass, weeds, “needles”, and other debris were removed so that the soil sampling area would be bare. The soil core was pushed as deep as possible into the soil but no deeper than 10 cm. Between trees, the core sampler was wiped clean and disinfected in 10% Clorox. All tools were either autoclaved or thoroughly cleaned with a 10% solution of Clorox. To eliminate DNA contamination of soil samples for ETOH preservation, nitrile gloves were worn, and between tree samples gloves were either cleaned with Clorox wipe or a new pair used.
Once the soil was collected into the labeled zip lock bag, the soil was immediately mixed by shaking the bag. Transferred from the bag using a sterilized metal spoon, small aliquots of soil were sieved through a ¼ in galvanized steel hardware cloth (screen) into an autoclaved paper cup. By folding the edge of the paper cup, small amounts of sieved soil were then poured into a pre-weighted and labeled 15 ml plastic ultra-high performance centrifuge tube containing 8 ml of 95% ETOH. Soil was added until a total volume of 11 ml was reached. To standardize conditions and maximize mixing, the tube was shaken frequently during the process. The tubes were immediately placed on ice and transported back to the lab. In the lab, the tubes were re-weighed to determine grams of field soil added to each tube. Afterward, sample tubes were stored in a freezer until shipment to LSU.
Also taken back to the lab was the leftover unsieved soil in the zip lock bag (approx. 80 ml), for soil-moisture determination (no need to refrigerate). In the lab, this soil was sieved with the same sieve type used for the centrifuge sample soil. Approximately 50 g of sieved field soil from each tree was placed in a weighed aluminum pan and dried at 105°C for at least 48 hrs. Dish was removed from the oven and allowed to cool. Once cooled, the dish was re-weighed and the difference from the pre-drying weight was calculated, which calculates the weight of the water in the original sample. This information was used to calculate soil moisture (dry wt. basis). Percent moisture content of soil is equal to weight of water in soil sample divided by the weight of dry soil.
Spiking Soil Methods: As a means to determine the suitability of the LSU soil sampling procedure to detect Ralstonia and various wetwood bacteria in a soil sample, 6 soil samples were spiked with ooze from slices of tissue from selected trees (positive for R. solanacearum species complex (RSSC) and Nasutitermes takasagoensis (Nt)). To determine the impact of soil on the detection of bacteria in the ooze, straight ooze was also sent. Similar to the soil collection procedures, precautions were taken to keep things clean. This included wearing Nitrile gloves and disinfecting with 10% Clorox solution or Clorox wipes.
Trees from which termite and soil samples were previously collected for LSU and which were positive for R. solanacearum species complex (RSSC) and Nasutitermes takasagoensis (Nt), were processed and ooze collected. In the field, slices of wood from the tree were collected and placed into disinfected plastic tubs containing sterile water and a few sheets of sterile paper towels (roughly 6). Water level was at least 1 cm deep and did not exceed ½ the thickness of the wood slice. To reduce evaporation, tubs were covered with lids or plastic wrap. To increase the odds of collecting an adequate amount of ooze, several slices were collected. Slices ranged in thickness, from 1 to 4 inches. Slices were collected mainly from trunks; however, large lower branches and/or roots were occasionally sampled. At the time of cutting, slices were scraped clean of most of the saw dust debris using a disinfected machete. The chain saw was cleaned between trees by operating in a bucket of water and then 10% Clorox solution for roughly 30 seconds. Slices were either placed in the lab or outside the lab on a table in the shade.
At the same time tree slice samples were collected, a soil sample consisting of 5 soil cores was collected in a zip-lock bag using the same procedure as outlined previously. This sample was kept out of direct sunlight and transported back to the lab, where it was stored on the lab bench until ooze was collected.
The tree slices were examined over the next 2 days. If total ooze production was deemed inadequate (less than 0.5 ml), slices might be left for additional time. In some cases, additional wood slices were collected from the same tree. In such a case, all the slices would be processed together after 72 hrs from when the first slices were collected. Ooze was collected using a vacuum pump fitted with a 1 ml pipet tip. The aspirate was collected in a centrifuge tube that had been cut to size and placed in the vacuum flask. Areas of ooze production on the cut sections were vacuumed until 3 ml of ooze was collected. If aspirate was less than 3 ml, drops of sterile distilled water were added to ooze production areas and the vacuuming process continued until 3 ml was reached. After ooze was collected, the presence of Rs was confirmed by testing the combined scrapings from all the pieces with an Agdia Rs-specific Immunostrip test. From the 3 ml of ooze suspension, a 0.06 ml subsample was removed and place it in a 0.5 ml Eppendorf tube containing 0.44 ml of 95% ETOH.
The soil sample that was set aside on the lab bench at the time the slices were collected was sieved using the same procedure as before (refer to soil collection methods). While avoiding contamination with skin cells, 10 g of sieved soil was placed into a new zip-lock bag. The remaining ooze suspension after taking the subsample (2.94 ml) was thoroughly mixed with the soil and then that spiked soil was poured into a pre-weighted and labeled 15 ml plastic ultra-high performance centrifuge tube containing 8 ml of 95% ETOH and stored in a freezer until shipment to LSU.
Brief Methods: Obj.3, Sub-obj. 3, Louisiana State University (termite analysis):
Methods were devised for meta-analysis to answer the hypothesized question: Are Nasutitermes takasagoensis, Coptotermes gestroi and Microcerotermes crassus, which represent the major termite species associated with IWTD, vectors for Ralstonia and other pathogenic bacteria causing IWTD?
To test this hypothesis, it was essential to collect termite samples in equal numbers that tested negative or positive for Ralstonia in Guam as this would service as the replicated set in order to (1) describe the bacterial taxa associated with workers attacking ironwood trees in Guam to test if termites carry plant pathogens, (2) test for relations between the tree-, plot-, and location-related factors associated with ironwood trees attacked by workers and microbial diversity of those worker samples, (3) determine if termites prefer feeding on parts of ironwood trees with low pathogen content compared to high pathogen content, and (4) determine whether R. solanacearum bacteria are ingested and survive in the termite gut. The Quantitative Insights into Microbial Ecology (QIIME2) pipeline (Caporaso et al. 2010, Estaki et al. 2020) version 2021-4, accessible on a server of the Hubbard Center for Genome Studies at the University of New Hampshire, was used to perform sequence data analysis. Demultiplexed sequencing reads were obtained from University of New Hampshire after Illumina sequencing in FASTQ format.
Forty-five termite samples were collected in 2019-20 by the University of Guam from healthy as well as sick ironwood trees present on 14 distinct locations on the island of Guam. Geology related (location, parent material classification, site management), tree-related (tree DS, tree health, presence or absence of Ralstonia, altitude classification) and plot-related (plot average DS, plot average health, stand maturity estimate, percentage of trees with termites in the plot, percentage of dead trees in the plot) parameters were recorded. The samples including soldiers and worker termites were partitioned into 70% ethanol (for morphological identification) and 95% ethanol (for Illumina sequencing) and were shipped to Louisiana State University. In the attached report by LSU, a full description of the materials and methods for the following topics can be found: (1) description of the factors collected from ironwood tree plots by University of Guam; (2) Morphological species identification of termites; (3) DNA extraction; (4) Primer selection; (5) DNA amplification and sequencing; (6) Bioinformatics analysis; (7) termite feeding experiments.
A more in-depth description of LSU’s methods for years 1-2 can be found in their progress report: LSU WSARE R&E Progress Report 4.1.21 - 3.30.22
A more in-depth description of LSU’s methods for year 3 can be found in the thesis of Garima Setia, LSU graduate student: https://digitalcommons.lsu.edu/gradschool_theses/5699/
Methods: Obj.3, Sub-obj. 3, Louisiana State University (soil analysis): A portion of each soil sample was first taken from the ethanol suspension and dried on a clean weighing boat. For each sample 250mg was used for DNA extraction with Qiagen DNeasy PowerSoil Pro Kit. Briefly, the samples were transferred into the PowerBead Pro Tubes with lysis buffer (CD 1) and homogenized by Benchmark Scientific BeadBlaster Tissue Homogenizer. The suspension was then processed and purified following the kit handbook. The DNA concentrations were quantified by Invitrogen Qubit 4 Fluorometer with dsDNA BR assay kit and 2.5 µl/ng DNA per sample was shipped to the University of New Hampshire for next-generation sequencing. The V1-V3 hyper variable region of the bacterial 16S rRNA gene was amplified from the DNA samples using one forward (27F) and two reverse primers (519Rmod and 519Rmodbio) to capture a broad range of biodiversity. QIIME2 was used to analyze the demultiplexed sequencing reads obtained from UNH after Illumina sequencing accessible on a server of their Hubbard Center for Genome Studies. Forward reads were subjected to further processing with removal of primers sequences and truncation to 251 nucleotides by DADA2. The rarefaction, alpha diversity (faith-pd, ASV richness, evenness and Shannon) and beta diversity (permanova and permdisp) were then conducted according to the tree-related factors and the visualization was completed by QIIME2 View and R package iNEXT. Taxonomical assignment was performed by comparing each sequence to the SILVA reference database.
Obj. 2: UH Res & UOG Res: Research into the bacterial wilt pathogen to determine the origin of Guam’s infection and its genomic biology.
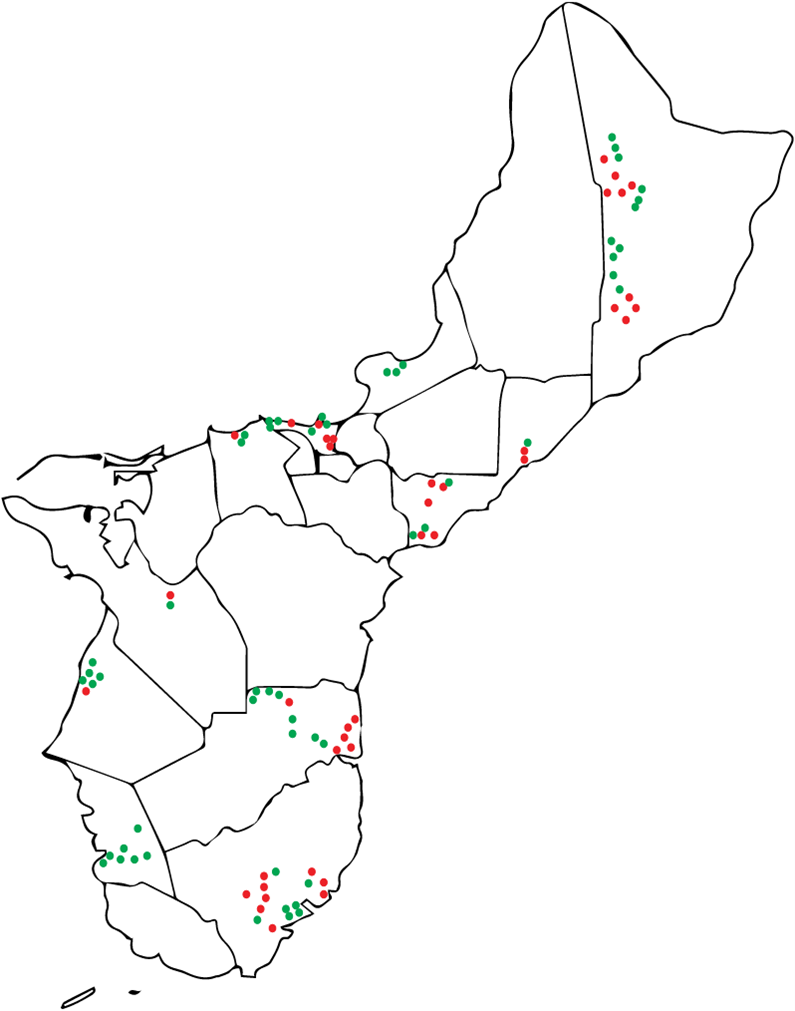
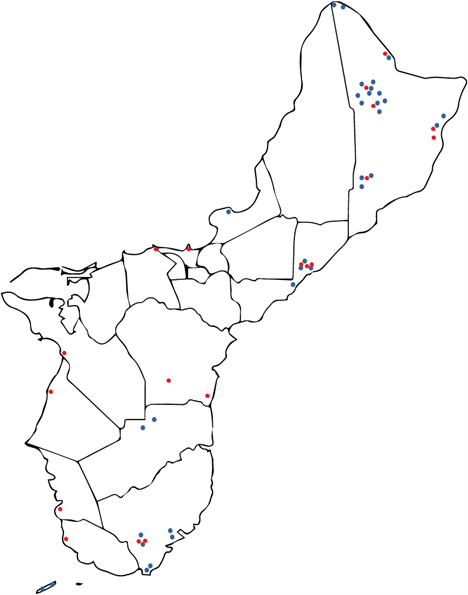
Results: Obj. 2, University of Guam: a total of 112 trees were sampled from 29 different locations in Guam. To determine the presence or absence of RSSC in the trees, drill shavings from each tree were tested with the RSSC specific Agdia immunostrips. Forty-five RSSC infected ironwood trees were identified. The map in Figure 5 shows the approximate locations of the sampled trees. Infected trees were widespread occurring in northern, central, and southeastern parts of the island. From visual inspection, trees in the southwestern area of the island appeared to less likely to test positive for RSSC.
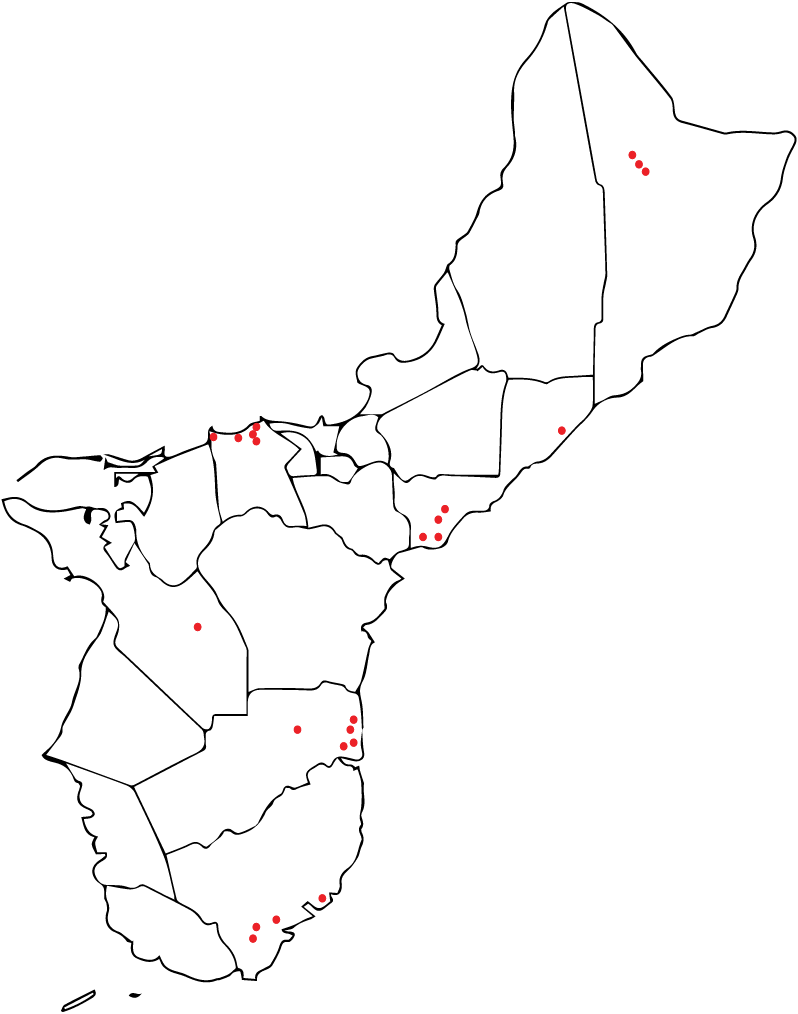
Three types of ooze were found in root cross-sections (VO = viscous ooze, WO = clear watery ooze, and MO = mixture of viscous ooze and watery ooze). The white to off-white viscous ooze (VO) mostly occurring in the outer portion of a root slice, nearly always tested positive for RSSC. This ooze gave a higher percentage of successful RSSC isolations than other ooze types. Of the 95 trees that tested positive for Rs only 35 were found to be suitable for collection of root slices and the production of ooze (Figure 6).
Thirty-five water cultures were prepared and sent to UH. The tree data from which the samples were collected is in Table 1.
Figure 5: Map of Guam with approximate locations of the ironwood trees sampled in the study. Trees that tested positives with Agdia immunostrip are in red, those that tested negative are in green.
Figure 6: Guam Ironwood tree locations where tissue samples were collected and Ralstonia solanacearum successfully isolated and sent to Hawaii.
Table 1: Underlying data on RSSC isolates obtained from Guam
|
UOG isolatea |
UOG Clinic entryb |
Ooze typec |
Height (m)d |
DBH (cm)e |
DSf |
Altitude (m)g |
Geologyh |
Sporocarpsi |
Termitesj |
|
19-124 |
19-124 |
mo |
19.2 |
26.51 |
3 |
71 |
0 |
0 |
1 |
|
19-127 |
19-127 |
mo |
12.5 |
32.69 |
4 |
78 |
0 |
0 |
0 |
|
19-135 |
19-135 |
vo |
128 |
25.79 |
3 |
176 |
0 |
1 |
2 |
|
19-147 |
19-147 |
mo |
19.6 |
17.63 |
3 |
176 |
0 |
1 |
2 |
|
19-156 |
19-156 |
vo |
15.5 |
9.39 |
4 |
26 |
0B |
0 |
1 |
|
19-157 |
19-157 |
vo |
9.8 |
23.56 |
3 |
19 |
0B |
0 |
1? |
|
19-158 |
19-158 |
vo |
8 |
19.74 |
2 |
21 |
0B |
0 |
0 |
|
19-161 |
19-161 |
vo |
12.8 |
31.52 |
1 |
117 |
1 |
0 |
0 |
|
19-170 |
19-170 |
mo |
13.8 |
75.15 |
4 |
-3 |
0B |
1 |
1 |
|
19-174 |
19-174 |
vo |
9.1 |
32.48 |
4 |
19 |
0B |
0 |
0(H) |
|
19-194 |
19-194 |
vo |
15.6 |
29.61 |
3 |
19 |
0B |
0 |
1 |
|
19-200 |
19-200 |
vo |
10.3 |
22.77 |
3 |
105 |
1 |
0 |
0 |
|
19-202 |
19-202 |
vo |
10 |
37.89 |
3 |
96 |
1 |
0 |
1H |
|
19-203 |
19-203 |
vo |
11.9 |
31.52 |
4 |
93 |
1 |
0 |
1A |
|
19-224 |
19-224 |
vo |
15.6 |
69.74 |
4 |
17 |
0B |
0 |
1H |
|
19-227 |
19-227 |
vo |
14.8 |
41.40 |
3 |
27 |
0B |
0 |
1 |
|
19-228 |
19-228 |
mo |
12.2 |
60.19 |
2 |
26 |
0B |
0 |
1 |
|
19-229 |
19-229 |
vo |
6.9 |
28.34 |
2 |
130 |
0 |
0 |
1H |
|
20-016 |
20-016 |
vo |
18 |
40.44 |
0 |
0 |
0 |
0 |
0 |
Table 1 Glossary: aUOG isolate, consisting of University of Guam clinic number with identifiers for multiple isolates; bUOG clinic number, identifies tree from with root samples were collected; cOoze type: vo = viscous ooze, mo = mixture of viscous ooze and watery ooze; dHeight, tree height; eDBH, tree diameter at breast height; fDS, decline severity, DS-0 (symptomless) to 4 (near dead); gAltitude, meters above sea level of tree site; hGeology, tree site parent material, 0= limestone upland, 0B= limestone beach, 1= volcanic upland, 1B=volcanic beach.
Results: Obj. 2, University of Hawaii: All the strains isolated from ironwood were able to grow on modified Kelman Tetrazolium Chloride (TZC) media and modified SMSA media. The growth was observed within 48 hours with light to dark pink pigmentation at 28oC. The identity of strains was first confirmed using colony PCR using Ralstonia solanacearum species complex (RSSC) specific end point PCR primers. The PCR products were electrophoresed at 100V for 40 minutes to visualize the amplicons. All the 25 strains isolated from ironwood were found to be positive with RSSC specific primers. Genomic DNA was extracted from all the strains using DNeasy Blood and Tissue kit (Qiagen, Valencia, CA). The extracted DNA was amplified using RSSC specific dnaA primers. All the strains were found to be positive. The BLASTn results showed all the strains from ironwood to be Ralstonia pseudosolanacearum.
Genome biology and evolution: Three sequence clusters (lineages) were defined by the hierarchical Bayesian analysis population structure method (BAPS) using 7 complete genomes of Ralstonia strains isolated from ironwood plus another 10 Ralstonia sp. strains from NCBI GenBank representing different phylotypes. The reconstructed genealogy of the 17 Ralstonia strains using ClonalFrameML revealed a ratio of recombination and mutation rates equal to R/θ = 0.09592, an average length of recombined fragments δ = 22.3743, a divergence of DNA imported by recombination ν = 0.160891 and a relative effect of recombination and mutation r/m = (R/θ) x δ x ν = 0.345. Moreover, each recombination event has been introduced on an average of δν = 3.59 substitutions. Four hundred fifty-five recombination events were identified on the branches of the clonal genealogy tree; out of these, 122 imports were observed in the ancestor branch from where both ironwood strains S28 and S26 of the phylotype II originated. This indicated that homologous recombination has played a significant role in the diversification/evolution of these strains. The most recombined strains were those from phylotype I within the lineage 3. Strain 19-170 appeared as the most recombinant one with 173 DNA imported fragments, following strains 19-228, S-14, S-5 and 19-200 positioned with 148, 135, 133 and 100 events. On the other hand, fastGEAR was used to identify ancient and recent recombination events across the core-genome of the 17 Ralstonia strains. Intriguingly, only one ancestral recombination event of 290 bp donated by the lineage 3 to lineage 2 seems to have occurred during the evolution time. Conversely, 572 recent recombination events were found across the population, which seems to indicate that these Ralstonia strains keep recombining possibly to adapt to a constraining environment. These 572 events were found to occur in 330 genes, which means that 2% of the total core-genome (1700 genes) have undergone recombination according to the Highways Enumerated by Recombination Observations (HERO) analysis. In agreement with ClonalFrameML results, the ironwood strains 19-170, 19-228, 19-200, S-14 and S-5 appeared as the highest recombinant strains based on the fastGEAR algorithm. However, strain CMR15 led the position with 209 recent events. No signals of recombination were detected on the ironwood strain S28 whereas just 2 events were found on the Ralstonia strains LLRS-1 from phylotype IV isolated from tobacco leaf veins. Surprisingly, 450 events were designated as external origin, meaning that these events have been imported by either other Ralstonia sp. or other closely related species sharing the same niche within the 17 Ralstonia isolates used in this study. Strains within the lineage 2 (phylotype IV) appeared as the main donors giving a total of 29 DNA fragments mainly to the strains of the lineage 3 (phylotype I), while lineage 1 (phylotype II) donated 25 events mainly to strains of the phylotype I. Lastly, the recombination network performed by HERO displayed a total of 195 pairs of recombining events between the three sequence clusters (lineages). One highway pair, which is defined as the donor-recipient pair that recombine more frequently with each other rather than with other lineages in the population, was found within strains of the same lineage 3. Nine events were detected within this one highway of recombination. This points that a low rate of homologous recombination is occurring between the strains of the same phylotype I, corroborating the fastGEAR output that the high rate of recombination inferred in the ironwood strains of phylotype I has been originated mainly from outside of the population (external origin) and just few events has been acquired from lineages 2 and 1 (phylotype IV and II, respectively).
A more in-depth description of the results for this objective can be found in the thesis of Sujan Paudel, UH graduate student: http://hdl.handle.net/10125/70339
Obj. 3: UH Res., LSU Res., & UOG Res.: Research into the flora of ironwood trees and the guts of termites.
Obj. 3 Sub-obj. 1: UH Res. & UOG Res.: Research into the fungal flora of ironwood trees and their likely role in IWTD.
Obj.3 Sub-obj. 2: UH Res. & UOG Res.: Research into the bacterial flora of ironwood trees and their likely role in IWTD.
Obj.3 Sub-obj. 3: LSU Res. & UOG Res.: Research to determine if termites carry ironwood bacteria and thus, might be responsible for their movement.
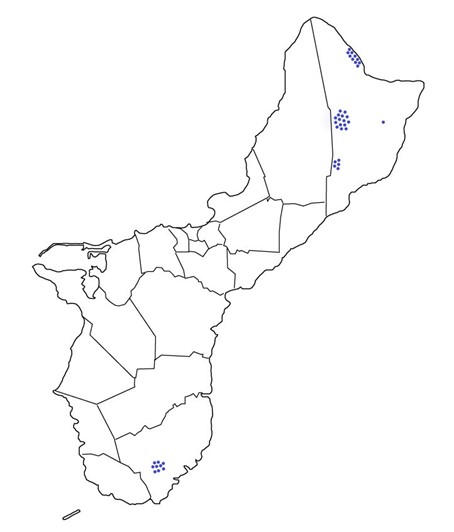
Results: Obj. 3, Sub-obj. 1 & Sub-obj. 2, University of Guam: UOG collected microbiome samples for Hawaii in four areas of the island (Figure 7). Tree information from the 40 ironwood trees from which microbiome samples were collected and sent to the University of Hawaii (Table 2).
Figure 7. Ironwood tree microbiome collection sites on Guam.
Table 2. University of Guam: Tree information from the 40 ironwood trees from which microbiome samples were collected and sent to the University of Hawaii.
| Ironwood Tree Study WSARE # SW19-906 | |||||||||||||||||||||||||||||||||
| Woody tissue Collection Sites for the University of Hawaii October/November 2021 | |||||||||||||||||||||||||||||||||
| Tree Tag No. | Tree GPS | Location | Tree DS | Tree Health Ranking | Human impact | Height (ft) | DBH Single [S] or Multiple [M] (cm) | Sporocarp | Site condition | Altitude (m) | Altitude Classification | Parent material | Parent material Classification | Termite activity | Suspected termite species | Rs (+/-) | Date of Rs Testing | Conductivity value µS/cm |
Conductivity level | Date Conductivity tissue collected | UOG Clinic no. assigned to shallow woody tissue sample | UOG Clinic no. assigned to deep woody tissue sample | Date of shallow and deep tissue sample collection | UOG Clinic no. assigned to root sample | Root Sample Rs (+/-) |
UOG Clinic no. assigned to 150 cm felled sample | 150cm Sample Rs(+/-) |
UOG Clinic no. assigned to 300 cm felled sample | 300cm Sample Rs(+/-) |
UOG Clinic no. assigned to 450 cm felled sample | 450cm Sample Rs (+/-) |
Date of Root sample and felled samples collection | Comments |
| 120 | 13.56690, 144.87727 | Watson's Farm, Yigo | 4 | 1 | 2 | 56.0 | 28.14 [S] | 1 | 1 | 165 | 2 | Limestone | 1 | 1 | N/A | (+) | 10/21/2021 | 750 | 2 | 11/3/21 | 21-267 | 21-268 | 11/3/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 121 | 13.56691, 144.87717 | Watson's Farm, Yigo | 4 | 1 | 2 | 51.0 | 29.11 [S] | 0 | 1 | 160 | 2 | Limestone | 1 | 2 | N/A | (+) | 10/21/2021 | 820 | 2 | 11/3/21 | 21-265 | 21-266 | 11/3/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 276 | 13.56555, 144.87750 | Watson's Farm, Yigo | 4 | 1 | 0 | 36 | 26.27 [S] | 0 | 1 | 164 | 2 | Limestone | 1 | 1 | N/A | (+) | 10/21/2021 | 753 | 2 | 11/3/21 | 21-263 | 21-264 | 11/3/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 277 | 13.56715, 144.87656 | Watson's Farm, Yigo | 4 | 1 | 0 | 31 | 21.02 [S] | 0 | 0 | 163 | 2 | Limestone | 1 | 2 | Nt | (+) | 10/21/2021 | 478 | 1 | 11/3/21 | 21-277 | 21-278 | 11/3/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | had to drill at an angle to get deep woody tissue sample due to tree being hollowed out |
| 279 | 13.56723, 144.87694 | Watson's Farm, Yigo | 2 | 1 | 0 | 59 | 24.66 [S] | 0 | 1 | 161 | 2 | Limestone | 1 | 2 | Nt | (+) | 10/22/2021 | 623 | 2 | 11/3/21 | 21-275 | 21-276 | 11/3/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 280 | 13.56694, 144.87735 | Watson's Farm, Yigo | 1 | 0 | 0 | 60 | 29.10 [S] | 0 | 0 | 159 | 2 | Limestone | 1 | 1 | N/A | (+) | 10/22/2021 | 858 | 3 | 11/3/21 | 21-269 | 21-270 | 11/3/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 282 | 13.56693, 144.87736 | Watson's Farm, Yigo | 1 | 0 | 0 | 45 | 31.53 [S] | 0 | 0 | 159 | 2 | Limestone | 1 | 1 | N/A | (+) | 10/22/2021 | 668 | 2 | 11/3/21 | 21-271 | 21-272 | 11/3/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 283 | 13.56708,144.87755 | Watson's Farm, Yigo | 2 | 1 | 0 | 61 | 47.70 [S] | 0 | 0 | 161 | 2 | Limestone | 1 | 2 | Nt | (+) | 10/22/2021 | 984 | 3 | 11/3/21 | 21-273 | 21-274 | 11/3/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 294 | 13.56715, 144.87604 | Watson's Farm, Yigo | 1 | 0 | 0 | 34 | 19.40 [S] | 0 | 1 | 167 | 2 | Limestone | 1 | 1 | N/A | (-) | 10/28/2021 | 243 | 0 | 11/3/21 | 21-279 | 21-280 | 11/3/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 275 | 13.56568, 144.87702 | Watson's Farm, Yigo | 4 | 1 | 1 | 34.0 | 25.87 [S] | 0 | 0 | 169 | 2 | Limestone | 1 | 2 | Nt | (+) | 10/21/2021 | 781 | 2 | 12/1/21 | 21-355 | 21-356 | 12/1/21 | 21-357 | + | 21-358 | + | 21-359 | + | 21-360 | + | 12/1/21 | had to drill at an angle to get deep woody tissue sample due to tree being hollowed out/ root sample taken at 1 inch depth due to small root diameter |
| 278 | 13.56717, 144.87664 | Watson's Farm, Yigo | 4 | 1 | 0 | 50 | 25.47 [S] | 0 | 0 | 169 | 2 | Limestone | 1 | 2 | Mc | (+) | 10/22/2021 | 582 | 2 | 12/1/21 | 21-343 | 21-344 | 12/1/21 | 21-345 | + | 21-346 | + | 21-347 | + | 21-348 | + | 12/1/21 | |
| 281 | 13.5669, 144.87734 | Watson's Farm, Yigo | 1 | 0 | 0 | 38 | 18.60 [S] | 0 | 0 | 169 | 2 | Limestone | 1 | 2 | C | (+) | 10/22/2021 | 559 | 1 | 12/1/21 | 21-361 | 21-362 | 12/1/21 | 21-363 | + | N/A | N/A | N/A | N/A | N/A | N/A | 12/1/21 | root sample taken at 1 inch depth due to small root diameter/ Rs sample taken from both roots and placed into the same container (all other samples were only taken from one root) |
| 290 | 13.56695, 144.87644 | Watson's Farm, Yigo | 2 | 1 | 0 | 44 | 30.47 [M] | 0 | 1 | 169 | 2 | Limestone | 1 | 2 | Nt | (-) | 10/21/2021 | 266 | 0 | 12/1/21 | 21-337 | 21-338 | 12/1/21 | 21-339 | - | 21-340 | - | 21-341 | - | 21-342 | - | 12/1/21 | small arboreal nest |
| 291 | 13.56695, 144.87663 | Watson's Farm, Yigo | 2 | 1 | 0 | 49 | 21.34 [S] | 0 | 1 | 169 | 2 | Limestone | 1 | 1 | N/A | (-) | 10/21/2021 | 384 | 1 | 12/1/21 | 21-349 | 21-350 | 12/1/21 | 21-351 | - | 21-352 | - | 21-353 | - | 21-354 | - | 12/1/21 | |
| 292 | 13.56715, 144.87598 | Watson's Farm, Yigo | 3 | 1 | 0 | 52 | 18.43 [S] | 0 | 1 | 169 | 2 | Limestone | 1 | 1 | N/A | (-) | 10/28/2021 | 412 | 1 | 12/1/21 | 21-331 | 21-332 | 12/1/21 | 21-333 | - | 21-334 | - | 21-335 | - | 21-336 | - | 12/1/21 | |
| 285 | 13.53233, 144.87451 | Yigo Experiment Station | 3 | 1 | 0 | 36 | 50.13 [S] | 0 | 1 | 116 | 2 | Limestone | 1 | 2 | Nt | (+) | 10/22/2021 | 455 | 1 | 11/22/21 | 21-323 | 21-324 | 11/22/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | had to drill at an angle to get deep woody tissue sample due to tree being hollowed out |
| 286 | 13.53233, 144.87451 | Yigo Experiment Station | 2 | 1 | 0 | 30 | 81.66 [S] | 0 | 1 | 116 | 2 | Limestone | 1 | 1 | N/A | (+) | 10/22/2021 | 970 | 3 | 11/22/21 | 21-325 | 21-326 | 11/22/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | had to drill at an angle to get deep woody tissue sample due to tree being hollowed out |
| 287 | 13.53233, 144.87451 | Yigo Experiment Station | 0 | 0 | 0 | 34 | 43.25 [S] | 1 | 1 | 116 | 2 | Limestone | 1 | 3 | Nt | (+) | 10/22/2021 | 715 | 2 | 11/22/21 | 21-321 | 21-322 | 11/22/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 288 | 13.53205, 144.874420 | Yigo Experiment Station | 4 | 1 | 0 | 30 | 31.53 [S] | 0 | 1 | 116 | 2 | Limestone | 1 | 1 | N/A | (+) | 10/22/2021 | 475 | 1 | 11/22/21 | 21-327 | 21-328 | 11/22/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 289 | 13.5309, 144.87422 | Yigo Experiment Station | 4 | 1 | 0 | 43 | 28.30 [S] | 0 | 0 | 116 | 2 | Limestone | 1 | 2 | Mc | (+) | 10/22/2021 | 496 | 1 | 11/22/21 | 21-329 | 21-330 | 11/22/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 218 | 13.26318, 144.71814 | Ija Experiment Station | 3 | 1 | 1 | 50 | 68.66 [M] | 1 | 0 | 74 | 1 | Residium | 2 | 2 | Nt | (+) | 10/13/2021 | 851 | 3 | 11/17/21 | 21-303 | 21-304 | 11/17/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 219 | 13.26335, 144.71809 | Ija Experiment Station | 3 | 1 | 0 | 26 | 39.78 [S] | 1 | 0 | 74 | 1 | Residium | 2 | 1 | N/A | (+) | 12/26/2019 | 850 | 3 | 11/17/21 | 21-305 | 21-306 | 11/17/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | Had to drill at an angle to get deep woody tissue sample due to tree being hollowed out. Extensive sporocarps |
| 258 | 13.26563, 144.71668 | Ija Experiment Station | 4 | 1 | 1 | 21 | 32.99 [S] | 1 | 1 | 85 | 1 | Residium | 2 | 1 | N/A | (+) | 10/13/2021 | 1257 | 3 | 11/17/21 | 21-311 | 21-312 | 11/17/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 259 | 13.26561, 144.71660 | Ija Experiment Station | 3 | 1 | 1 | 30 | 47.78 [M] | 1 | 1 | 85 | 1 | Residium | 2 | 1 | N/A | (-) | 10/13/2021 | 800 | 2 | 11/17/21 | 21-307 | 21-308 | 11/17/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 260 | 13.26544, 144.71675 | Ija Experiment Station | 2 | 1 | 0 | 29 | 43.66 [S] | 0 | 2 | 84 | 1 | Residium | 2 | 3 | Nt | (-) | 10/13/2021 | 583 | 2 | 11/17/21 | 21-313 | 21-314 | 11/17/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 261 | 13.26526, 144.71742 | Ija Experiment Station | 1 | 0 | 0 | 26 | 19.40 [S] | 0 | 1 | 96 | 1 | Residium | 2 | 0 | N/A | (-) | 10/13/2021 | 338 | 1 | 11/17/21 | 21-315 | 21-316 | 11/17/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | Had to drill at an angle to get deep woody tissue sample due to tree being hollowed out. |
| 262 | 13.26571, 144.71752 | Ija Experiment Station | 3 | 1 | 0 | 30 | 33.95 [S] | 0 | 0 | 92 | 1 | Residium | 2 | 2 | Ct | (-) | 10/13/2021 | 302 | 1 | 11/17/21 | 21-317 | 21-318 | 11/17/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 263 | 13.26629, 144.71758 | Ija Experiment Station | 1 | 0 | 1 | 62 | 48.51 [S] | 1 | 1 | 90 | 1 | Residium | 2 | 3 | Nt | (-) | 10/13/2021 | 538 | 1 | 11/17/21 | 21-319 | 21-320 | 11/17/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | Extensive sporocarps |
| 264 | 13.26558, 144.71663 | Ija Experiment Station | 4 | 1 | 1 | 34 | 28.14 [S] | 0 | 1 | 85 | 1 | Residium | 2 | 1 | N/A | (-) | 10/13/2021 | 608 | 2 | 11/17/21 | 21-309 | 21-310 | 11/17/21 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 265 | 13.62520, 144.89521 | Tarague Beach, AAFB | 4 | 1 | 1 | 63.0 | 33.64 [M] | 0 | 1 | 16 | 1 | Coral Sand | 3 | 1 | N/A | (-) | 10/14/2021 | 762 | 2 | 10/14/21 | 21-293 | 21-294 | 11/10/2021 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 266 | 13.62525, 144.89513 | Tarague Beach, AAFB | 3 | 1 | 1 | 51.0 | 53.82 [M] | 0 | 1 | 18 | 1 | Coral Sand | 3 | 1 | N/A | (-) | 10/14/2021 | 530 | 1 | 10/14/21 | 21-287 | 21-288 | 11/10/2021 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 267 | 13.62535, 144.89515 | Tarague Beach, AAFB | 3 | 1 | 1 | 53.0 | 29.10 [S] | 0 | 1 | 16 | 1 | Coral Sand | 3 | 1 | N/A | (-) | 10/14/2021 | 285 | 0 | 10/14/21 | 21-289 | 21-290 | 11/10/2021 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 268 | 13.62533, 144.89516 | Tarague Beach, AAFB | 3 | 1 | 1 | 39.0 | 24.25 [S] | 0 | 1 | 16 | 1 | Coral Sand | 3 | 1 | N/A | (-) | 10/14/2021 | 377 | 1 | 10/14/21 | 21-291 | 21-292 | 11/10/2021 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 269 | 13.62536, 144.89515 | Tarague Beach, AAFB | 1 | 0 | 1 | 61.0 | 30.25 [M] | 0 | 1 | 15 | 1 | Coral Sand | 3 | 1 | N/A | (-) | 10/14/2021 | 475 | 1 | 10/14/21 | 21-285 | 21-286 | 11/10/2021 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 270 | 13.62543, 144.89495 | Tarague Beach, AAFB | 1 | 0 | 1 | 62.0 | 75.67 [S] | 0 | 1 | 17 | 1 | Coral Sand | 3 | 1 | N/A | (-) | 10/14/2021 | 311 | 1 | 10/14/21 | 21-283 | 21-284 | 11/10/2021 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 271 | 13.62528, 144.89466 | Tarague Beach, AAFB | 0 | 0 | 1 | 67 | 80.04 [S] | 0 | 1 | 17 | 1 | Coral Sand | 3 | 1 | N/A | (-) | 10/14/2021 | 214 | 0 | 10/14/21 | 21-281 | 21-282 | 11/10/2021 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 272 | 13.62456, 144.89565 | Tarague Beach, AAFB | 0 | 0 | 1 | 36 | 74.70 [S] | 0 | 1 | 30 | 1 | Coral Sand | 3 | 3 | Nt | (-) | 10/14/2021 | 308 | 1 | 10/14/21 | 21-295 | 21-296 | 11/10/2021 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 273 | 13.62460, 144.89586 | Tarague Beach, AAFB | 0 | 0 | 1 | 35 | 93.96 [S] | 0 | 1 | 25 | 1 | Coral Sand | 3 | 1 | N/A | (-) | 10/14/2021 | 225 | 0 | 10/14/21 | 21-297 | 21-298 | 11/10/2021 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| 274 | 13.62428, 144.89581 | Tarague Beach, AAFB | 1 | 0 | 1 | 55.0 | 61.05 [M] | 0 | 1 | 32 | 1 | Coral Sand | 3 | 1 | N/A | (-) | 10/15/2021 | 254 | 0 | 11/15/21 | 21-299 | 21-300 | 11/10/2021 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | Had to drill at an angle to get deep woody tissue sample due to tree being hollowed out. |
| 231 | 13.56938, 144.93198 | Andersen Golf Course | 4 | 1 | 1 | 35 | 56.86[M] | 1 | 2 | 162 | 2 | Limestone | 1 | 1 | N/A | (+) | 10/15/2021 | 500 | 1 | 11/10/21 | 21-301 | 21-302 | 11/10/2021 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
Glossary found here: FINAL trees for Hawaii microbiome_for report
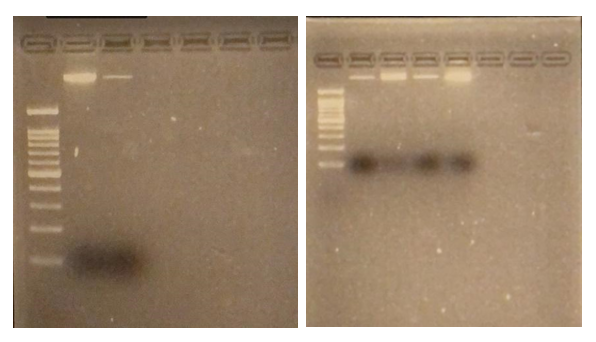
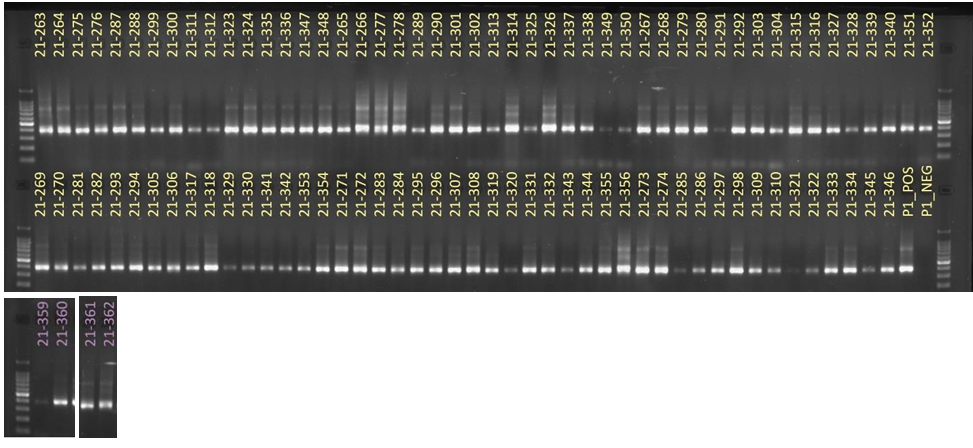
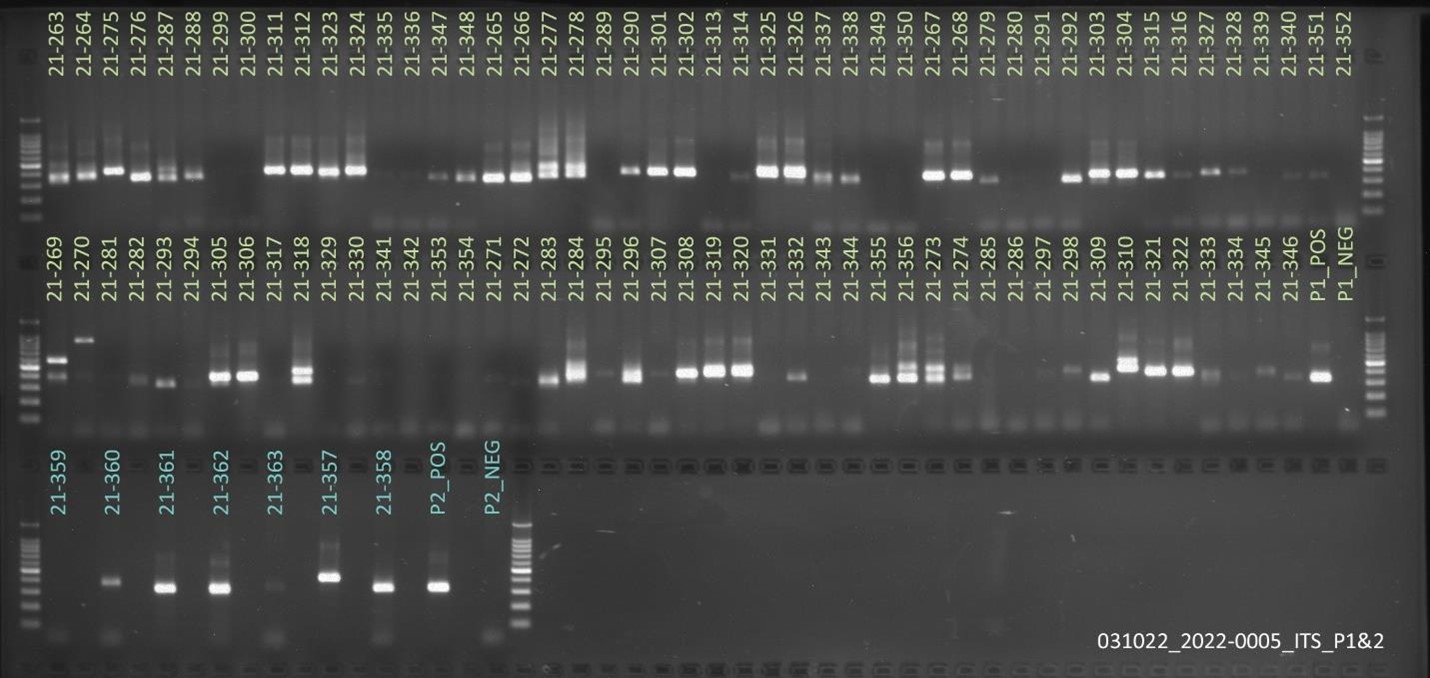
Results: Obj 3, Sub-obj. 2, University of Hawaii: Modified protocol for DNA isolation from greenhouse ironwood plants worked for greenhouse ironwood plants and was repeated 5 times to confirm the results. The obtained results were consistent (Figure 8). Left figure shows three band columns: standards, greenhouse ironwood, ironwood tree. Right figure shows five band columns: standards, ironwood tree, greenhouse ironwood, another ironwood tree and greenhouse ironwood.
Hawaii received approximately 101 samples from Guam. The samples’ 16S rRNA amplified gene region can be seen in Figure 9.
For ITS sequencing, the gel picture showing amplification within 70 samples out of 101 can be seen in Figure 10.
In microbiome analyses, Proteobacteria and Basidiomycota were the dominant bacterial and fungal phyla identified among the samples. A high number of ITS reads remained unclassified, probably due to the poorly annotated fungal database. In bacterial community analyses, Ralstonia was identified with high accuracy in infected ironwood samples. Lower bacterial richness was obtained with the samples infected with Ralstonia.
Figure 8. Results of DNA isolation from greenhouse ironwood plants.
Figure 9. 16S rRNA amplified gene region of samples from Guam.
Figure 10. ITS sequencing results of 70 out of the 101 samples sent from Guam.
A more in-depth description of the results for this objective can be found in the thesis of Diksha Klair, UH graduate student: https://hdl.handle.net/10125/103907
Results: Obj. 3, Sub-obj. 3, University Of Guam:
Termite Results: from 2019-2021, the University of Guam collected termites from infested ironwood trees across Guam. In 2019 and 2021, a total of 45 termite samples from 44 ironwood trees were collected and sent to Louisiana State University for species identification and analysis of gut contents (Table 3 and Figure 11, blue dots). In 2021, an additional 35 termite samples from 30 ironwood trees were collected and sent to Louisiana State University for species identification and analysis of gut contents (Table 3 and Figure 11, red dots).
Soil Results: in 2022, the University of Guam collected soil from the same ironwood trees that the termites had been collected from. A total of 73 soil samples were collected from 68 trees and sent to Louisiana State University for further analysis (Table 3 and Figure 11, blue and red dots).
Spiked Soil Results: also in 2022, the University of Guam collected bacterial ooze and additional soil from 6 of the ironwood trees in the study. The bacterial ooze was used to spike a total of 6 soil samples, which were sent to LSU to determine the suitability of the soil sampling procedure to detect Ralstonia and various wetwood bacteria in a soil sample.
Table 3. University of Guam: Tree information from ironwood trees from which termites and soil were collected and sent to Louisiana State University.
| Ironwood Tree Study WSARE # SW19-906 | ||||||||||||||||||||||||||||||
| LSU Termite Study (Nt and Cg) and Soil Study 2019-2022 | ||||||||||||||||||||||||||||||
| UOG Clinic no. assigned to termites | Termite Collection Date | Termite ID (morphological) | Source of Termites | Tree Tag No. | Tree GPS | Location | Tree Alive or Dead | Tree DS | Tree Health Ranking | Tree Condition | Height (m) | DBH Single [S] or Multiple [M] (cm) | Sporocarp | Site condition | Altitude (m) | Altitude Classification | Parent material | Parent material Classification | Rs Testing Date | Rs (+/-) | Soil Cover | UOG Clinic no. assigned to soil sample | Depth of soil cores taken | Soil Tube # | % Soil Moisture | Conductivity Value | Conductivity Level | Soil Sample and Conductivity Collection Date | Candidate for ooze collection? | Comments |
| 19-76 | 06-19-19 | Nt | tree | 165 | 13.53216, 144.87418 | UOG Yigo Station | 0 | 3 | 1 | 1 | 9.0 | 18.57 [M] | 0 | 1 | 148 | 2 | limestone | 1 | 02-05-21 | (+) | needles&ferns | 22-130 | 9,10,8,10,5 | 9 | 30.24% | 367 | 1 | 2/23/22 | NO | Tree now dead (2/23/22), conk now present |
| 19-77 | 06-20-19 | Nt | tree | 166 | 13.56583, 144.87688 | Watson's Farm, Yigo | 0 | 3 | 1 | 1 | 11.1 | 33.60 [S] | 0 | 1 | 170 | 2 | limestone | 1 | 02-05-21 | (+) | needles&sm trees | 22-141 | 8,10,10,10,10 | 20 | 33.83% | 493 | 1 | 2/24/2022 | Root | |
| 19-78 | 06-20-19 | Nt | tree | 167 | 13.56598, 144.87462 | Watson's Farm, Yigo | 0 | 4 | 1 | 1 | 10.0 | 9.16 [M] | 0 | 1 | 164 | 2 | limestone | 1 | 02-05-21 | (-) | needles&ferns | 22-140 | 10,10,10,10,10 | 19 | 31.20% | 185.4 | 0 | 2/24/2022 | Trunk | |
| 19-79 | 06-20-19 | Nt | tree | 168 | 13.56706, 144.87537 | Watson's Farm, Yigo | 0 | 2 | 1 | 2 | 13.0 | 24.20 [M] | 1 | 1 | 171 | 2 | limestone | 1 | 02-18-21 | (-) | needles&sm trees | 22-138 | 10,8,10,6,4 | 17 | 46.67% | 415 | 1 | 2/24/2022 | Branch, Root | |
| 19-80 | 06-20-19 | Nt | tree | 169 | 13.56660, 144.87416 | Watson's Farm, Yigo | 0 | 0 | 0 | 1 | 13.7 | 22.13 [S] | 0 | 1 | 161 | 2 | limestone | 1 | 02-18-21 | (-) | needles&ferns | 22-139 | 8,10,10,6,10 | 18 | 40.84% | 192.7 | 0 | 2/24/2022 | NO | |
| 19-81 | 06-20-19 | Nt | tree | 170 | 13.56456, 144.87746 | Watson's Farm, Yigo | 0 | 1 | 0 | 1 | 8.2 | 13.69 [S] | 0 | 1 | 168 | 2 | limestone | 1 | 02-18-21 | (-) | needles&ferns | 22-137 | 10,6,8,10,4 | 16 | 34.46% | 267 | 0 | 2/24/2022 | Trunk | |
| 19-82 | 06-21-19 | Nt | tree | 171 | 13.26583, 144.71691 | UOG Ija Station | 0 | 3 | 1 | 1 | 12.6 | 26.27 [S] | 0 | 1 | 96 | 1 | residum | 2 | 03-16-21 | (+) | grass | 22-147 | 10,10,10,10,10 | 26 | 48.82% | 482 | 1 | 3/2/2022 | Trunk | |
| 19-83 | 06-21-19 | Nt | tree | 172 | 13.26529, 144.71629 | UOG Ija Station | 0 | 4 | 1 | 1 | 17.2 | 33.76 [S] | 1 | 1 | 92 | 1 | residum | 2 | 03-16-21 | (-) | needles&weeds | 22-151 | 10,10,10,10,10 | 30 | 38.28% | 378 | 1 | 3/2/2022 | Root | |
| 19-84 | 06-27-19 | Nt | tree | 173 | 13.47111, 144.84528 | Mangilao Golf Course | 0 | 0 | 0 | 1 | 9.2 | 17.61 [M] | 0 | 2 | 129 | 2 | limestone | 1 | 03-02-21 | (-) | needles&ferns | 22-126 | 10,10,10,10,10 | 5 | 16.26% | 202 | 0 | 2/17/2022 | ||
| 19-85 | 06-27-19 | Nt | tree | 174 | 13.47082, 144.84503 | Mangilao Golf Course | 0 | 4 | 1 | 1 | 9.2 | 39.17 [S] | 0 | 2 | 127 | 2 | limestone | 1 | 03-02-21 | (+) | needles | 22-133 | 10,10,10,10,10 | 12 | 39.19% | 370 | 1 | 2/23/22 | Branch | Conk now present |
| 19-86 | 07-01-19 | Nt | tree | 176 | 13.37520, 144.73877 | Windward Golf Course | 0 | 4 | 1 | 2 | 12.0 | 33.28 [S] | 1 | 2 | 117 | 2 | residium | 2 | 03-16-21 | (-) | grass | 22-163 | 10,10,10,10,10 | 42 | 59.14% | 737 | 2 | 3/10/2022 | NO | Level 1 sieve was used to sieve soil |
| 19-87 | 07-01-19 | Nt | tree | 175 | 13.37788, 144.74162 | Windward Golf Course | 0 | 1 | 0 | 2 | 10.7 | 18.47 [S] | 1 | 2 | 126 | 2 | residium | 2 | 03-16-21 | (+) | grass | 22-164 | 6,10,8,8,10 | 43 | 35.97% | 429 | 1 | 3/10/2022 | NO | |
| 19-88 | 07-09-19 | Nt | tree | 177 | 13.64889, 144.85289 | Ritidian | 0 | 1 | 0 | 1 | 8.0 | 39.50 [S] | 0 | 1 | 12 | 1 | coral sand | 3 | 02-19-21 | (-) | needles&palm leaf debris | 22-187 | 10,10,10,10,10 | 55 | 17.69% | 361 | 1 | 3/17/2022 | NO | |
| 19-89 | 07-09-19 | Nt | tree | 163 | 13.65222, 144.85764 | Ritidian | 0 | 0 | 0 | 1 | 10.5 | 46.70 [S] | 0 | 1 | 21 | 1 | coral sand | 3 | 02-19-21 | (-) | needles | 22-286 | 10,10,10,10,10 | 54 | 9.31% | 241 | 0 | 3/17/2022 | NO | |
| 19-90 | 07-12-19 | Nt | tree | 178 | 13.23476, 144.64574 | Cocos Island | 0 | 2 | 1 | 1 | 11.9 | 23.90 [S] | 0 | 0 | 20 | 1 | coral sand | 3 | 04-08-21 | (-) | needles | 22-208 | 10,4,8,10,10 | 66 | 24.59% | 227 | 0 | 3/24/2022 | NO | |
| 19-91 | 07-12-19 | Nt | tree | 179 | 13.23701, 144.65102 | Cocos Island | 0 | 1 | 0 | 1 | 10.3 | 17.80 [M] | 0 | 0 | 12 | 1 | coral sand | 3 | 04-08-21 | (-) | needles&large rocks | 22-209 | 2,4,10,4,10,6 | 67 | 24.98% | 212 | 0 | 3/24/2022 | NO | |
| 19-92 | 07-16-19 | Nt | tree | 180 | 13.46753, 144.84236 | Mangilao Golf Course | 0 | 1 | 0 | 1 | 6.9 | 15.50 [M] | 0 | 1 | 124 | 2 | limestone | 1 | 03-02-21 | (+) | needles | 22-124 | 10,10,10,10,10 | 3 | 39.21% | 277 | 0 | 2/17/2022 | NO | |
| 19-93 | 07-16-19 | Nt | tree | 181 | 13.46464, 144.85080 | Thousand Steps | 0 | 0 | 0 | 1 | 5.9 | 28.18 [S] | 0 | 0 | 17 | 1 | limestone | 1 | 03-24-21 | (-) | needles | 22-224 | 4,40,4,2,2,4,2 | 72 | 29.28% | 247 | 0 | 3/30/22 | NO | |
| 19-94 | 07-17-19 | Nt | tree | 182 | 13.24797, 144.72690 | Ysrael Beach | 0 | 4 | 1 | 1 | 9.4 | 24.20 [S] | 0 | 0 | 5 | 1 | coral sand | 3 | 03-19-21 | (-) | needles&grass | 22-170 | 4,10,10,10,10 | 49 | 11.23% | 930 | 3 | 3/10/2022 | NO | |
| 19-95 | 07-17-19 | Nt | tree | 183 | 13.24741, 144.72708 | Ysrael Beach | 0 | 1 | 0 | 1 | 5.0 | 32.64 [S] | 0 | 0 | 7 | 1 | coral sand | 3 | 03-19-21 | (-) | needles | 22-171 | 8,10,8,8,4 | 50 | 22.46% | 230 | 0 | 3/10/2022 | NO | |
| 19-96 | 07-18-19 | Nt | tree | 229 | 13.50311, 144.78416 | Sagan Kotturan Chamoru | 0 | 1 | 0 | 1 | 11.9 | 33.92 [S] | 0 | 0 | 45 | 1 | limestone | 1 | 03-26-21 | (-) | needles&weeds | 22-223 | 10,10,10,10,8 | 71 | 8.80% | 190.1 | 0 | 3/30/22 | NO | |
| 19-98 | 07-30-19 | Nt | tree | 185 | 13.56830, 144.93179 | AAFB | 0 | 3 | 1 | 1 | 10.2 | 36.46 [M] | 0 | 2 | 164 | 2 | limestone | 1 | 03-23-21 | (-) | grass&weeds | 22-156 | 10,10,10,10,9.5 | 35 | 56.23% | 205 | 0 | 3/3/2022 | NO | DS changed to level 4 as of 3-3-2022 |
| 19-99 | 07-30-19 | Nt | tree | 186 | 13.56314, 144.93079 | AAFB | 0 | 1 | 0 | 1 | 9.1 | 40.70 [M] | 0 | 1 | 160 | 2 | limestone | 1 | 03-23-21 | (-) | trees&weeds | 22-155 | 4,8,10,8,10 | 34 | 28.31% | 197 | 0 | 3/3/2022 | Root, Branch | |
| 19-100 | 08-09-19 | Nt | tree | 187 | 13.25978, 144.73688 | Duenas Beach | 0 | 0 | 0 | 2 | 12.4 | 63.70 [S] | 0 | 1 | 11 | 1 | coral sand | 3 | 03-18-21 | (-) | grass&needles | 22-168 | 10,10,10,10,10 | 47 | 7.24% | 264 | 0 | 3/10/2022 | NO | |
| 19-101 | 08-09-19 | Nt | tree | 188 | 13.25963, 144.73735 | Duenas Beach | 0 | 1 | 0 | 1 | 11.4 | 41.20 [S] | 0 | 1 | 12 | 1 | coral sand | 3 | 03-18-21 | (-) | needles&trees | 22-169 | 10,10,10,10,10 | 48 | 15.08% | 326 | 1 | 3/10/2022 | NO | |
| 19-102 | 08-20-19 | Nt | tree | 191 | 13.62340, 144.89664 | Tarague Beach | 0 | 0 | 0 | 1 | 11.6 | 39.00 [S] | 0 | 2 | 21 | 1 | coral sand | 3 | 03-26-21 | (-) | needles | 22-174 | 10,10,10,10,8 | 53 | 6.36% | 224 | 0 | 3/11/2022 | No | |
| 19-103 | 09-13-19 | Nt | tree | 199 | 13.56594, 144.87816 | Watson's Farm, Yigo | 0 | 1 | 0 | 1 | 12.1 | 30.90 [S] | 0 | 0 | 168 | 2 | limestone | 1 | 02-12-21 | (-) | needles&ferns | 22-190 | 2,4,4,4,4,8 | 58 | 39.51% | 227 | 0 | 3/17/2022 | NO | |
| 19-104 | 09-13-19 | Nt | tree | 200 | 13.56674, 144.87776 | Watson's Farm, Yigo | 0 | 0 | 0 | 1 | 8.1 | 22.30 [S] | 0 | 0 | 169 | 2 | limestone | 1 | 02-12-21 | (-) | needles&ferns | 22-146 | 10,4,8,10,10 | 25 | 54.61% | 164 | 0 | 2/24/2022 | Root | |
| 19-105 | 09-13-19 | Nt | tree | 76 | 13.56864, 144.87701 | Watson's Farm, Yigo | 0 | 1 | 0 | 1 | 9.2 | 26.27 [S] | 0 | 0 | 173 | 2 | limestone | 1 | 02-18-21 | (-) | needles&ferns | 22-189 | 4,4,6,6,6,10 | 57 | 43.11% | 259 | 0 | 3/17/2022 | NO | |
| 19-106 | 09-13-19 | Nt | tree | 85 | 13.56741, 144.87413 | Watson's Farm, Yigo | 0 | 1 | 0 | 1 | 7.3 | 23.10 [S] | 0 | 0 | 163 | 2 | limestone | 1 | 02-18-21 | (-) | needles&ferns | 22-188 | 4,6,6,8,10 | 56 | 53.90% | 224 | 1 | 3/17/2022 | NO | |
| 19-107 | 09-13-19 | Nt | tree | 86 | 13.56707, 144.87654 | Watson's Farm, Yigo | 0 | 0 | 0 | 1 | 12.4 | 37.70 [S] | 0 | 0 | 171 | 2 | limestone | 1 | 02-12-21 | (-) | needles&ferns | 22-143 | 8,8,10,8,4 | 22 | 49.40% | 225 | 0 | 2/24/2022 | Branch | |
| 19-108 | 09-13-19 | Nt | tree | 87 | 13.53309, 144.87161 | UOG Yigo Station | 0 | 0 | 0 | 1 | 19.7 | 33.30 [S] | 0 | 1 | 178 | 2 | limestone | 1 | 02-04-21 | (-) | needles&ferns | 22-127 | 10,10,10,10,10 | 6 | 34.33% | 217 | 0 | 2/23/22 | Root | |
| 19-109 | 09-13-19 | Nt | tree | 88 | 13.53297, 144.87364 | UOG Yigo Station | 0 | 1 | 0 | 0 | 8.9 | 25.20 [S] | 0 | 1 | 142 | 2 | limestone | 1 | 02-04-21 | (-) | grass&needles | 22-129 | 10,10,10,10,10 | 8 | 29.87% | 185.1 | 0 | 2/23/22 | Root | |
| 20-122 | 07-15-20 | Nt | under bark | 111 | 13.56711, 144.87700 | Watson's Farm, Yigo | 0 | 4 | 1 | 2 | 7 | 24.52 [S] | 1 | 1 | 168 | 2 | limestone | 1 | 07-15-20 | (+) | needles&grass | 22-142 | 10,10,10,6,10 | 21 | 42.34% | 804 | 2 | 2/24/2022 | Root | |
| 20-123 | 07-15-20 | Nt | under bark | 119 | 13.56692, 144.87740 | Watson's Farm, Yigo | 0 | 3 | 1 | 2 | 19.6 | 17.64 [M] | 0 | 1 | 163 | 2 | limestone | 1 | 07-15-20 | (+) | needles&ferns | 22-145 | 10,10,10,8,10 | 24 | 38.84% | 520 | 1 | 2/24/2022 | Trunk, Root | |
| 20-124 | 07-15-20 | Nt | mud tunnel | 112 | 13.56736, 144.87631 | Watson's Farm, Yigo | 0 | 3 | 1 | 2 | 7.4 | 13.38 [S] | 0 | 1 | 158 | 2 | limestone | 1 | 07-15-20 | (+) | ferns | 22-144 | 8,10,10,10,8 | 23 | 38.63% | 412 | 1 | 2/24/2022 | Root | |
| 20-125 | 08-01-20 | Nt | arboreal nest | 122 | 13.43109, 144.80084 | UOG, Mangilao | 0 | 4 | 1 | 0 | 15.1 | 17.52 [M] | 0 | 0 | 67 | 1 | limestone | 1 | 08-01-20 | (+) | needles&ferns | 22-122 | 4,7,4,10,10,4 | 2 | 25.57% | 759 | 2 | 2/11/2022 | Trunk | |
| 20-126 | 08-01-20 | Nt | under bark | 153 | 13.43128, 144.80281 | UOG, Mangilao | 0 | 3 | 1 | 2 | 11.9 | 41.40 [S] | 0 | 2 | 81 | 1 | limestone | 1 | 08-01-20 | (+) | grass | 22-121 | 10,9,6,10,10 | 1 | 27.69% | 598 | 2 | 2/11/2022 | ||
| 20-129 | 08-01-20 | Nt | under bark | 226 | 13.46781, 144.84233 | Mangilao Golf Course | 0 | 1 | 1 | 2 | 7.8 | 27.71 [S] | 0 | 2 | 129 | 2 | limestone | 1 | 08-01-20 | (+) | needles | 22-125 | 10,10,10,10,10 | 4 | 38.69% | 581 | 2 | 2/17/2022 | ||
| 20-130 | 08-01-20 | Nt | mud tunnel | 217 | 13.53231, 144.87267 | UOG Yigo Station | 0 | 4 | 1 | 2 | 13.9 | 31.21 [S] | 0 | 1 | 173 | 2 | limestone | 1 | 08-01-20 | (+) | needles&ferns | 22-131 | 10,10,10,10,10 | 10 | 38.99% | 713 | 2 | 2/23/22 | Root | Conk now present |
| 20-131 | 08-01-20 | Nt | mud tunnel | 217 | 13.53231, 144.87267 | UOG Yigo Station | 0 | 4 | 1 | 2 | 13.9 | 31.21 [S] | 0 | 1 | 173 | 2 | limestone | 1 | 08-01-20 | (+) | needles&ferns | 22-131 | 10,10,10,10,10 | 10 | 38.99% | 713 | 2 | 2/23/22 | Root | Conk now present |
| 20-132 | 08-02-20 | Nt | mud tunnel | 215* | 13.26533, 144.71634 | UOG Ija Station | 0 | 4 | 1 | 1 | 10 | 37.90 [S] | 0 | 1 | 96 | 1 | residum | 2 | 08-02-20 | (+) | sm trees&needles | 22-152 | 10,10,10,10,10 | 31 | 43.44% | 344 | 1 | 3/2/2022 | Root | |
| 20-133 | 08-02-20 | Nt | under bark | 216** | 13.26578, 144.71692 | UOG Ija Station | 0 | 4 | 1 | 2 | 11.9 | 31.53 [S] | 0 | 1 | 93 | 1 | residum | 2 | 08-02-20 | (+) | grass | 22-148 | 10,10,10,10,10 | 27 | 47.30% | 403 | 1 | 3/2/2022 | Trunk, Root | |
| 20-134 | 08-02-20 | Nt | under bark | 208 | 13.26590, 144.71707 | UOG Ija Station | 0 | 4 | 1 | 2 | 15.8 | 35.67 [S] | 1 | 1 | 82 | 2 | residum | 2 | 08-02-20 | (+) | grass | 22-150 | 10,10,10,10,10 | 28 | 45.75% | 639 | 2 | 3/2/2022 | Branch, Root | |
| 20-135 | 08-02-20 | Nt | arboreal nest | 209 | 13.26595, 144.71707 | UOG Ija Station | 0 | 4 | 1 | 2 | 8.9 | 29.78 [M] | 0 | 1 | 110 | 2 | residum | 2 | 08-02-20 | (+) | grass | 22-149 | 10,10,10,10,10 | 29 | 50.52% | 449 | 1 | 3/2/2022 | Trunk | Branch cut to evaluate ooze on 03/02/2022 |
| 21-150 | 9/17/2021 | Cg | tree | 306 | 13.28276, 144.75675 | UOG Inarajan Station | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | 16 | 1 | limestone | 1 | n/a | n/a | needles&trees | 22-167 | 4,10,10,8,10 | 46 | 31.63% | n/a | n/a | 3/10/2022 | NO | STUMP only, no tree |
| 21-151 | 9/17/2021 | Cg | tree | 306 | 13.28276, 144.75675 | UOG Inarajan Station | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | 16 | 1 | limestone | 1 | n/a | n/a | needles&trees | 22-167 | 4,10,10,8,10 | 46 | 31.63% | n/a | n/a | 3/10/2022 | NO | STUMP only, no tree |
| 21-152 | 9/17/2021 | Mc | tree | n/a | 13.28300, 144.75600 | UOG Inarajan Station | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
| 21-153 | 9/17/2021 | Cg | stake | 302 | 13.26530, 144.71624 | UOG Ija Station | 1 | n/a | n/a | 1 | 7.6 | 29.77 [M] | 0 | 1 | 89 | 1 | residum | 2 | n/a | n/a | leaves&sm trees | 22-153 | 10,10,10,10,10 | 32 | 37.03% | n/a | n/a | 3/2/2022 | NO | |
| 21-154 | 9/17/2021 | Cg | tree | 302 | 13.26530, 144.71624 | UOG Ija Station | 1 | n/a | n/a | 1 | 7.6 | 29.77 [M] | 0 | 1 | 89 | 1 | residum | 2 | n/a | n/a | leaves&sm trees | 22-153 | 10,10,10,10,10 | 32 | 37.03% | 351 | 1 | 3/2/2022 | NO | |
| 21-155 | 9/17/2021 | Cg | tree | 216 | 13.26578, 144.71692 | UOG Ija Station | 0 | 4 | 1 | 2 | 11.9 | 31.53 [S] | 0 | 1 | 93 | 1 | residum | 2 | 08-02-20 | (+) | grass | 22-148 | 10,10,10,10,10 | 27 | 47.30% | 403 | 1 | 3/2/2022 | Trunk, Root | |
| 21-156 | 9/17/2021 | Cg | tree | 301 | 13.36490, 144.64920 | Nimitz Park | 0 | 0 | 0 | 0 | 13.1 | 75.99 [S] | 0 | 2 | 4 | 1 | coral sand | 3 | 3/4/2022 | (-) | grass | 22-162 | 10,4,6,8,8 | 41 | 26.29% | 300 | 0 | 3/4/2022 | Root | |
| 21-157 | 9/17/2021 | Cg | tree | 378 | 13.47956, 144.75429 | Paseo Park, Hagatna | 0 | 0 | 0 | 0 | 9.4 | 60.70 [M] | 0 | 2 | 6 | 1 | limestone | 1 | 3/4/2022 | (-) | grass | 22-159 | 4,4,4,8,4 | 38 | 7.51% | 232 | 0 | 3/4/2022 | NO | Soil collected from only 1/4 of circumference of tree, remaining 3/4 circumference was fully exposed to beach & water (no soil) |
| 21-159 | 9/17/2021 | Cg | tree | 250 | 13.47040, 144.84463 | Mangilao Golf Course | 1 | n/a | n/a | 2 | 6.7 | 17.35 [S] | 1 | 1 | 128 | 2 | limestone | 1 | n/a | n/a | needles | 22-132 | 10,4,10,10,10 | 11 | 36.84% | 802 | 2 | 2/23/2022 | No | Tree is now dead (2/23/22) |
| 21-160 | 9/22/2021 | Cg | tree | 32 | 13.40239, 144.66307 | Apaca Point, Agat | 0 | 0 | 0 | 1 | 13.1 | 103.13 [M] | 1 | 1 | 6 | 1 | residum | 2 | 3/4/2022 | (-) | needles&weeds | 22-161 | 10,10,10,10,10 | 40 | 52.65% | 237 | 0 | 3/4/2022 | NO | |
| 21-161 | 9/22/2021 | Cg | tree | 32 | 13.40239, 144.66307 | Apaca Point, Agat | 0 | 0 | 0 | 1 | 13.1 | 103.13 [M] | 1 | 1 | 6 | 1 | residum | 2 | 3/4/2022 | (-) | needles&weeds | 22-161 | 10,10,10,10,10 | 40 | 52.65% | 237 | 0 | 3/4/2022 | NO | |
| 21-162 | 9/22/2021 | Cg | tree | 379 | 13.47870, 144.73200 | Governor's Complex, Aniquia | 0 | 0 | 0 | 2 | 10.9 | 50.13 [M] | 0 | 2 | 6 | 1 | residum | 2 | 3/4/2022 | (-) | needles | 22-160 | 6,10,6,10,10 | 39 | 17.76% | 172.8 | 0 | 3/4/2022 | Branch | |
| 21-163 | 9/22/2021 | Mc | tree | 222 | 13.477254, 144.75693 | Padre Palmo Park, East Agana | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
| 21-164 | 9/22/2021 | Cg | tree | 154 | 13.53327, 144.87158 | UOG Yigo Station | 0 | 4 | 1 | 1 | 38.4 | 33.96 [S] | 1 | 2 | 144 | 2 | limestone | 1 | 2/23/2022 | (+) | weeds&needles | 22-128 | 10,10,10,10,10 | 7 | 40.00% | 384 | 1 | 2/23/22 | Trnk, Brnch, Root | Ganoderma present 2/23/22 |
| 21-165 | 9/22/2021 | Cg | tree | 297 | 13.56553, 144.87749 | Watson's Farm, Yigo | 0 | 4 | 1 | 1 | 33.0 | 93.98 [S] | 1 | 1 | 159 | 2 | limestone | 1 | 3/17/2022 | (+) | needles&leaves | 22-191 | 10,10,10,10,6 | 59 | 20.74% | 1,113 | 3 | 3/17/2022 | Root | |
| 21-166 | 9/22/2021 | Cg | tree | 296 | 13.56528, 144.87704 | Watson's Farm, Yigo | 0 | 3 | 1 | 1 | 12.8 | 41.2 [S] | 0 | 1 | 159 | 2 | limestone | 1 | 3/17/2022 | (+) | needles&grass&ferns | 22-192 | 6,8,10,10,10 | 60 | 31.58% | 444 | 1 | 3/17/2022 | Root | |
| 21-168 | 9/27/2021 | Cg | tree | 240 | 13.55782, 144.93009 | AAFB | 0 | 4 | 1 | 2 | 12.5 | 55.36 [M] | 0 | 2 | 155 | 2 | limestone | 1 | 3/3/2022 | (-) | needles | 22-158 | 10,10,10,10,6 | 37 | 38.51% | 308 | 1 | 3/3/2022 | NO | 3rd trunk is dead (cut off). Still measured for DBH [M] |
| 21-170 | 9/27/2021 | Cg | tree | 242 | 13.56132, 144.93056 | AAFB | 0 | 1 | 0 | 0 | 13.1 | 83.17 [M] | 0 | 1 | 155 | 2 | limestone | 1 | 3/3/2022 | (-) | noni&needles&ferns | 22-157 | 8,10,8,10,10 | 36 | 54.76% | 395 | 1 | 3/3/2022 | Root | |
| 21-171 | 9/27/2021 | Cg | tree | 243 | 13.62518, 144.89525 | AAFB, tarague beach | 0 | 4 | 1 | 2 | 13.1 | 47.7 [S] | 0 | 2 | 9 | 1 | coral sand | 3 | 3/11/2022 | (+) | needles&grass | 22-173 | 10,10,10,10,10 | 52 | 7.87% | 703 | 2 | 3/11/2022 | No | |
| 21-172 | 9/27/2021 | Cg | tree | 244 | 13.62629, 144.89449 | AAFB, tarague beach | 0 | 3 | 1 | 0 | 16.2 | 50.94 [S] | 0 | 1 | 9 | 1 | coral sand | 3 | 3/11/2022 | (-) | needles | 22-172 | 10,10,10,10,10 | 51 | 10.89% | 521 | 1 | 3/11/2022 | No | |
| 21-174 | 9/27/2021 | Cg | tree | 246 | 13.37650, 144.73789 | Windward hills golf course | 0 | 4 | 1 | 2 | 11.3 | 46.42 [M] | 0 | 2 | 115 | 2 | residum | 2 | 3/10/2022 | (+) | grass | 22-165 | 8,8,10,6,4 | 44 | 36.79% | 264 | 0 | 3/10/2022 | Root, Branch | |
| 21-175 | 9/27/2021 | Cg | tree | n/a | n/a | Mangilao Golf Course | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | 115 | 2 | limestone | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
| 21-176 | 9/29/2021 | Cg | tree | 250 | 13.47040, 144.84463 | Mangilao Golf Course | 1 | n/a | n/a | 2 | 6.7 | 17.35 [S] | 1 | 1 | 128 | 2 | limestone | 1 | n/a | n/a | needles | 22-132 | 10,4,10,10,10 | 11 | 36.84% | n/a | n/a | 2/23/2022 | No | Tree is now dead (2/23/22) |
| 21-177 | 9/29/2021 | Mc | tree | 249 | 13.47025, 144.84740 | Mangilao Golf Course | 0 | 2 | 1 | 1 | n/a | 32.34 [S] | 0 | 1 | 132 | 2 | limestone | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
| 21-178 | 9/29/2021 | Mc | tree | 247 | 13.46830, 144.84820 | Mangilao Golf Course | 0 | 3 | 1 | 0 | n/a | 23.45 [S] | 0 | 1 | 132 | 2 | limestone | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
| 21-179 | 9/29/2021 | Mc | tree | 248 | 13.46830, 144.84820 | Mangilao Golf Course | 1 | n/a | n/a | 1 | n/a | 40.83 [S] | 1 | 1 | 132 | 2 | limestone | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
| 21-180 | 9/29/2021 | Cg | tree | 251 | 13.37163, 144.76817 | Country Club of the Pacific | 0 | 3 | 1 | 2 | 9.4 | 54.17 [S] | 0 | 2 | 26 | 1 | residum | 2 | 3/10/2022 | (-) | grass | 22-166 | 4,8,6,8,8,4 | 45 | 17.59% | 298 | 0 | 3/10/2022 | No | 6 soil cores collected instead of 5 to get enough soil |
| 21-181 | 9/29/2021 | Cg | tree | 253 | 13.26586, 144.71689 | UOG Ija Station | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | 75 | 1 | residum | 2 | n/a | n/a | grass | 22-217 | 10,10,10,10,10 | 68 | 42.74% | n/a | n/a | 3/29/2022 | No | STUMP only, no tree; soil sample not processed immediately but back at lab |
| 21-182 | 9/29/2021 | Cg | tree | 254 | 13.26566, 144.71705 | UOG Ija Station | 0 | 3 | 1 | 0 | 10.4 | 30.72 [S] | 0 | 1 | 86 | 1 | residum | 2 | 3/2/2022 | (+) | weeds, grass | 22-154 | 10,10,10,10,10 | 33 | 56.83% | 584 | 2 | 3/2/2022 | Branch | Large branch off of trunk |
| 21-183 | 9/29/2021 | Cg | tree | 303 | 13.26587, 144.71694 | UOG Ija Station | 1 | n/a | 1 | 1 | 28.0 | 45.9[M] | 0 | 2 | 86 | 1 | residum | 2 | n/a | n/a | grass | 22-195 | 10,10,10,10,10 | 63 | 41.90% | n/a | n/a | 3/18/22 | No | |
| 21-184 | 9/29/2021 | Cg | tree | 304 | 13.26592, 144.71698 | UOG Ija Station | 1 | n/a | 1 | 1 | 23.0 | 39.24[M] | 1 | 2 | 86 | 1 | residum | 2 | n/a | n/a | grass | 22-196 | 10,10,10,10,10 | 64 | 37.10% | n/a | n/a | 3/18/22 | no | |
| 21-185 | 9/29/2021 | Cg | tree | 305 | 13.26597, 144.71705 | UOG Ija Station | 1 | n/a | 1 | 1 | 28.0 | 40.42[S] | 0 | 2 | 86 | 1 | residum | 2 | n/a | n/a | grass | 22-197 | 8,10,10,10,10 | 65 | 38.20% | n/a | n/a | 3/18/22 | No | |
| 21-186 | 9/29/2021 | Cg | tree | 255 | 13.27161, 144.66414 | Merizo Cemetary | 0 | 3 | 1 | 0 | 40.0 | 29.10[S] | 0 | 0 | 25 | 1 | residum | 2 | 3/18/22 | (+) | weeds,shrubs,needles | 22-194 | 10,10,10,10,8 | 62 | 32.68% | 540 | 1 | 3/18/22 | No | |
| 21-187 | 9/29/2021 | Mc | tree | 256 | 13.29400, 144.66210 | Fort Soledad | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
| 21-188 | 9/29/2021 | Cg | tree | 257 | 13.29509, 144.66092 | Fort Soledad | 0 | 3 | 1 | 2 | 35.0 | 47.7[S] | 0 | 2 | 42 | 1 | residum | 2 | 3/18/22 | (-) | grass | 22-193 | 10,10,8,4,4 | 61 | 26.93% | 442 | 1 | 3/18/22 | No | |
| *Tree 215 was the source tree for two Ralstonia solanacearum species complex isolates used by Sujan Paudel in this thesis research at the University of Hawaii in August 2020. https://scholarspace.manoa.hawaii.edu/handle/10125/70339; Paudel hawii 0085O 10777.pdf The corresponding UOG isolate numbers are 192021 and 19202 and the associated UOG clinic number is 19-202. |
||||||||||||||||||||
| **Tree 216 was the source tree for one Ralstonia solanacearum species complex isolate used by Sujan Paudel in this thesis research at the University of Hawaii in August 2020. https://scholarspace.manoa.hawaii.edu/handle/10125/70339; Paudel hawii 0085O 10777.pdf The corresponding UOG isolate number is 19203 and the associated UOG clinic number is 19-203. |
Full Glossary and Table, as well as additional sub-tables with soil moisture and bacterial ooze data, can be found here: FINAL_LSU Ironwood termite and soil metadata 1-11-23
Figure 11. Ironwood tree termite collection sites on Guam. Blue dot = 2019 collection site; Red dot = 2021 collection site.
Brief Results: Obj.3, Sub-obj. 3, Louisiana State University (termite analysis): Based on a collection of termites from 48 ironwood trees, the ‘higher’ termite, Nasutitermes takasagoensis was the most common accounting for 93% of the samples. The lack of Ralstonia spp. and the low presence and abundance of wetwood bacteria in the gut of these termites suggest that they are not a vector for these pathogens of IWTD. At taxonomic level two in SILVA (Phylum level), Spirochaetes (48.22 %) and Fibrobacteres (41.43 %) were found to be the most dominant phyla followed by Bacteroidetes (3.61%), Proteobacteria (3.35%), Margulisbacteria (0.84%), Acidobacteria (0.77%), Planctomycetes (0.65%), and others (1.61%). It was determined that the presence or absence of Ralstonia in trees from which termites were collected showed no significant effect on the total bacteria community; however, the communities of termites collected from sick trees showed significantly higher bacterial richness compared to those from healthy trees. In the attached report by LSU, a full description of the results can be found.
A more in-depth description of LSU’s resutls for years 1-2 can be found in their progress report: LSU WSARE R&E Progress Report 4.1.21 - 3.30.22
A more in-depth description of LSU’s results for year 3 can be found in the thesis of Garima Setia, LSU graduate student: https://digitalcommons.lsu.edu/gradschool_theses/5699/
Results: Obj.3, Sub-obj. 3, Louisiana State University (soil analysis): No Ralstonia spp.were observed among the sequences obtained from the 40 soil samples. However, Ralstonia was detected in the positive controls (soil spiked with Ralstonia), which is evidence that the methods for Ralstonia detection were working. Firmicutes (50.67%) and Actinobacteria (31.52%) were found the most abundant phyla in soil followed by Proteobacteria (10.57%), Acidobacteria (3.96%) and Planctomycetes (0.77%). Tree-and plot related factors (such as altitude, percentage of dead trees, and percentage of trees with termites) showed no significant effects of on any of the alpha diversity indices. Altitude and parent material classification had significant effects on differentiation of soil bacteria communities (beta-diversity). Further analyses including comparison between the microbiota of termites and soil samples are currently under way.
A great deal of research was accomplished during the reporting period of this project; however, in the coming months more will come to light as theses, meeting abstracts, and journal articles are published. From the analysis of 26 water cultures of the bacterial wilt pathogen by the University of Hawaii, it was determined that the bacterial wilt pathogen in Guam’s trees is best described as Ralstonia solanacearum species complex (RSSC). Their analysis revealed two strains and thus two potential origins of Guam’s pathogen. The most common was Phylotype I which is present in North and Eastern Asia, whereas less common was Phylotype II which occurs in Central America. Based a collection of termites from 48 ironwood trees, the ‘higher’ termite, Nasutitermes takasagoensis was the most common accounting for 93% of the samples. The lack of Ralstonia spp. and the low presence and abundance of wetwood bacteria in the gut of these termites suggests that they are not a vector for these pathogens of IWTD. At taxonomic level two in SILVA (Phylum level), Spirochaetes (48.22 %) and Fibrobacteres (41.43 %) were found to be the most dominant phyla followed by Bacteroidetes (3.61%), Proteobacteria (3.35%), Margulisbacteria (0.84%), Acidobacteria (0.77%), Planctomycetes (0.65%), and others (1.61%). It was determined that the presence or absence of Ralstonia in trees from which termites were collected showed no significant effect on the total bacterial community; however, the communities of termites collected from sick trees showed significantly higher bacterial richness compared to those from healthy trees. When the research findings of this project are applied to the large concept of forest health, several relationships between healthy trees (healthy forest) and unhealthy trees (unhealthy forest) emerge. These relationships offer opportunities by which selected environmental sampling and analyses can be used to ascertain the health of trees and thus the forest.
Research outcomes
This project arose from the need to address questions from farmers, landowners, and government agencies seeking answers and solutions to why there are thousands of ‘sick’ ironwood trees (Casuarina equisetifolia) on thousands of acres across Guam and what can be done about it. Prior to the decline and death of ironwood trees in 2002 (IWTD), recommendations for sustainable agriculture production on Guam always included the planting of ironwood trees from the Guam Department of Agriculture. Ironwood is one of the dominant, naturally occurring and propagated agroforestry species in the Pacific due to its ability to withstand salt spray, typhoon strength winds, and poor soil conditions. Natural and planted stands of ironwood trees are critical for controlling erosion along coastlines, estuaries, and riverbanks. Those on the hillsides and along rivers reduce soil runoff into Guam’s coral reefs. The Guam Department of Agriculture ceased its yearly 25,000 ironwood seedling give-away program in large part due to IWTD.
As a result of research activities to understand the impact of bacterial wilt and bacterial wetwood on Guam’s ironwood tree, steps have been identified that will aid in restoring Casuarina equisetifolia as an agroforestry species of choice on the island of Guam. Knowing that most of the bacteria-infected trees on Guam and the neighboring islands occur in cohorts of a few trees gives disease assessment teams the tools to quickly identify infected trees and locations where tree removal would be most effective in reducing the impact of bacterial wilt in particular and IWTD in general. The concern that termites might spreading Ralstonia solanacearum and other IWTD pathogens was eliminated when research showed that none of the termite species Nasutitermes takasagoensis, Coptotermes gestroi and Microcerotermes crassus that attack ironwood trees in Guam were vectors for IWTD pathogens. To increase the gene pool of Guam’s trees through cross-pollination, hundreds of trees from eighteen C. equisetifolia seed lots from the Commonwealth Scientific and Industrial Research Organization (CSIRO) Australian Tree Seed Center (ATSC) have been planted during the course of this project. Some of these trees have been used to establish new ironwood windbreaks and others have been used as replacement trees in windbreaks with high levels of decline. Professionals and the general public are now being advised to reduce lawnmower and weed-trimmer damage to roots and the base of trees as a means to reduce infection and spread of pathogens. To reduce transmission of RSSC and wetwood bacteria when pruning, individuals are instructed to disinfect all tools. The public is also being advised to remove severely declined trees as a means to protect nearby healthy trees.
Based on what we now know about IWTD, we believe planting healthy young trees is the quickest way to replace trees in decline. Any young healthy ironwood tree will do, it can be from seed of a Guam tree or one of the cultivars from ATSC. The tree can be planted near a declining tree, provided the distance is great enough to prevent any commingling of their roots, 100 ft should be sufficient. The new tree can be planted in the same spot where a tree died, provided there are no tree remnants present such as pieces of trunk, branches, or roots. Since termites were found not to vector RSSC or any other likely bacterial causal agent, there is currently no reason to adjust the planting of ironwood in a location based on the presence or absence of termites.
The research from this project and related research of PI Robert Schlub has raised several concerns and offers direction for future research. Though R. solanacearum occurs in Guam, China, and India, it only accounts for 65% of the trees with IWTD symptoms in Guam, whereas in China and India it accounts for 100% of trees with bacterial wilt symptoms. Guam doesn’t have one Ralstonia solanacearum species complex (RSSC) but two. The most common was Phylotype I which is present in North and Eastern Asia, whereas less common was Phylotype II which occurs in Central America. RSSC is not limited to Guam’s ironwood trees but in 2022 was identified on the islands of Saipan, Rota, and Palau. Additionally, with the detection of RSSC in hosts other than ironwood in Guam, Saipan, Rota, Palau, and Tinian the threat posed by this RSSC will likely increase in the future. Having discovered that samples infected with R. solanacearum showed significantly lower bacterial richness and diversity provides evidence that deciphering the endophytic microbiota and knowing the role of associated biotic and abiotic factors will be advantageous in the application of bioformulations in the development of efficient disease management strategies. This project confirmed previous research findings that termite activity increases with IWTD severity; however, it failed to determine the actual role played by termites. The association between termites and IWTD might also be indirect. The mechanical tree damage due to termite feeding might be serving as a point of entry for the IWTD-associated pathogens. To understand the indirect association of termites with IWTD, the pattern of termite infestation at the different stages of IWTD severity needs to be studied. This would elucidate if the association of termites is causal to IWTD or if these termites simply serve as opportunistic feeders. At our current level of knowledge, nearly 35% of ironwood trees in decline cannot be explained by known biotic or abiotic factors. With continued research, new factors linked to IWTD will likely be discovered, and the role played by previously discovered agents will be refined. Determining the factors behind the cause and transmission of IWTD would help in developing an integrated disease management plan to prevent the spread of this decline to healthy ironwood trees within the island of Guam and the neighboring islands.
Education and Outreach
Participation summary:
Obj 1 Yr 1-2: UOG Ed: Through tree plantings, educate the public on the importance of planting new off-island cultivars (seedlots from international provenance trails)
Ironwood (Casuarina equisetifolia) seeds sourced from different geographical locations (Table 4) were planted into seed cones. Saplings were raised for 4-6 months then transplanted to different locations across Guam in February 2020 (Figure 12). Saplings were used for both new and replacement windrows.
Obj. 1, Sub-obj. 1, UOG: Plant trees of mixed cultivars in 10 visually deteriorating windrows across Guam.
Ten deteriorating windrows were identified by visual surveillance of sites for IWTD. These windrows were refurbished with 6-15 trees each, for a total of 138 trees planted (Table 5). Property owners were educated on the best tree care and management practices during planting.
Obj. 1, Sub-obj. 2, UOG: Construct 15 new ironwood agroforestry projects using 150 trees consisting of a mixture of tree cultivars.
Fifteen new windrows were created consisting of 10 trees each, for a total of 150 trees (Figure 13). Property owners were educated on the best tree care and management practices during planting.
Figure 12: Locations of windrows planted across Guam.
Figure 13: Extension Associate Joseph Afaisen planting a new windrow at Cooperator Vincent Velasquez’s farm. The rebar and fencing is used to protect the sapling from wild pigs.
Table 4: Locations of Casuarina equisetifolia seeds used for the purpose of planting new windrows and increasing genetic diversity.
| Location | State | Latitude | Longitude | Altitude (m) | |||||
| DEG | MIN | DEG | MIN | ||||||
| (R)BAKO NATIONAL PARK | MALAYSIA | 1 | 44 | 110 | 30 | 30 | |||
| (R)KUANTAN PENANG | MALAYSIA | 3 | 48 | 103 | 20 | 0 | |||
| (R)DESARU JOHOR | MALAYSIA | 1 | 30 | 104 | 16 | 0 | |||
| HORNO IS. MANUS | PAPUA NEW GUINEA | 2 | 19 | 147 | 49 | 1 | |||
| (R)ELA BEACH | PAPUA NEW GUINEA | 9 | 5 | 147 | 17 | 10 | |||
| (R)KOLOMBANGARA | SOLOMON ISLANDS | 8 | 7 | 57 | 8 | 2 | |||
| (R)BAN KAM PHUAM RANO | THAILAND | 9 | 21 | 98 | 27 | 10 | |||
| (R)HADSAMIRA SONGKHLA | THAILAND | 7 | 9 | 100 | 37 | 2 | |||
| MELVILLE ISLAND | AUSTRALIA | 11 | 5 | 130 | 40 | 10 | |||
| (R)AGAMY | EGYPT | 31 | 13 | 29 | 45 | 9 | |||
| (R)CHANDIPUR BALASORE | INDIA | 21 | 30 | 86 | 54 | 2 | |||
| HAMBANTOTA | SRIL | 6 | 8 | 81 | 7 | 16 | |||
| VILLACLARA PLANTED | CUBA | 23 | 0 | 80 | 30 | 60 | |||
| (R)BAOBAB | KENYA | 4 | 0 | 39 | 6 | 25 | |||
| (R)EFATE | VANUATU | 17 | 45 | 168 | 18 | 30 | |||
| (R)YANJING | CHINA | 23 | 0 | 113 | 3 | 4 | |||
| (R)DAODONG SF | CHINA | 19 | 58 | 110 | 59 | 10 | |||
| BEIHAI GUANGXI | CHINA | 21 | 35 | 109 | 0 | 2 | |||
| (R)NINH TUAN | VIETNAM | 11 | 33 | 108 | 59 |
2 |
|||
Table 5. Deteriorating windrows refurbished across Guam.
|
Location |
Date planted |
Number of trees planted |
Tree origins used |
GPS- beginning of windrow |
GPS- end of windrow |
|
Palm Tree Golf Course, AAFB |
June 2019 |
12 |
Guam (9), Vanuatu (3) |
13.57476, 144.93803 |
13.56912, 144.93175 |
|
Palm Tree Golf Course, AAFB |
June 2019 |
6 |
Guam (2), Vanuatu (2), China (2) |
13.56127, 144.93317 |
13.56187, 144.93282 |
|
Bernard Watson Farm, Yigo |
June 2019 |
15 |
Vanuatu (3), Malaysia (3), Egypt (2), PNG (1), China (3), Vietnam (1), Sri Lanka (2) |
13.56637, 144.87721
|
13.56702, 144.87700 |
|
Bernard Watson Farm, Yigo |
June 2019 |
15 |
Vanuatu (3), Malaysia (3), Egypt (2), PNG (1), China (3), Vietnam (1), Sri Lanka (2) |
13.56587 144.87730 |
13.56583, 144.87688
|
|
UOG Yigo Experiment Station |
August 2020 |
15 |
PNG (2), Thailand (4), Egypt (4), India (4), Malaysia (1) |
13.5335 144.8716 |
13.5335, 144.8710 |
|
UOG Yigo Experiment Station |
August 2020 |
15 |
Thailand (4), Egypt (4), India (5), China (2) |
13.5324 144.8745 |
13.5320, 144.8745 |
|
Mangilao Golf Course |
August 2020 |
15 |
Egypt (2), Thailand (5), Guam (4), India (2), Kenya (1), China (1) |
13.4684, 144.8423 |
13.4680, 144.8423 |
|
Mangilao Golf Course |
August 2020 |
15 |
Thailand (9), Guam (4), Australia (2) |
13.4680, 144.8423 |
13.4676, 144.8424 |
|
Windward Hills Golf Course |
August 2020 |
15 |
Egypt (2), Thailand (4), Guam (4), India (4), PNG (1) |
13.3754, 144.7480 |
13.3759, 144.7380 |
|
Windward Hills Golf Course |
August 2020 |
15 |
Thailand (4), Guam (4), India (4), PNG (1), Australia (2) |
13.3759, 144.7379 |
13.3765, 144.7379 |
Obj. 4 Yr 2-3: UOG Ed: Restoring ironwood as an agroforestry species in Guam through awareness and action of the local and scientific communities.
Obj. 4, Sub-obj. 1, UOG: Through two ½ day workshops, attendees will learn about the care of the ironwood trees and its uses in agroforestry. Ironwood trees will be given away.
½ Day Workshop February 28, 2020:
A hands-on half-day workshop/training was held for 11 University of Guam Pest Management students in the afternoon on Saturday February 28, 2020 (Figure 14). In an effort to bring attention to the current studies and familiarize the students with ironwood tree decline, participants were exposed to a variety of practices and techniques used in studying these phenomena.
Activity 1: Windbreak planting and genetic diversity: Participants were provided different ironwood trees sapling varieties from around the world and were tasked to refill the missing ironwood trees in a windbreak located at the University of Guam Agriculture and Life Science Building. Participants were also instructed on the importance of increasing the genetic diversity of Guam’s ironwood trees. All students successfully planted one tree.
Activity 2: Ironwood tree seed germination: Ironwood trees seeds from different areas of the world were given to the participants. Each participant was tasked to plant the seeds in cone planters filled with damp #4 Sunshine mix. Dr. Schlub demonstrated the proper depth the seeds should be buried. Each participant had seeds from different geographical locations and 20 cones and successfully planted the seeds. The saplings from the seeds will be maintained and used in planting windbreaks during the next rainy season.
Activity 3: Evaluating ironwood trees and testing for Ralstonia solanacearum (Rs): Participants were divided into three groups and taught how to visually determine ironwood tree decline severity. They logged the geographical location and altitude of a tree using a GPS device, determined the height of a tree using a Precision Laser Rangefinder with Hypsometer, and measured the circumference of the tree at breast height to calculate the diameter. Also, through visual inspection of the tree they determined if the tree had present or past evidence of termite activity as well as if there were Ganoderma sporocarps. Finally, they used a drill to collect wood shavings from the tree and tested for the presence of Rs in the laboratory using Agdia immunostrip test kits.
Activity 4. Inoculation of tomato seedlings with Ralstonia solanacearum (Rs) previously isolated from ironwood trees: Each participant inoculated tomato seedlings with Rs isolated from ironwood trees and grown on media. Some plants were lightly scraped on the stem near the crown and Rs was applied using a cotton swab. Some plants were lightly scraped with nothing applied. Half of the plants inoculated with Rs developed severe bacterial wilt symptoms, while no symptoms developed on the plants that weren’t inoculated. Photos of the plants were taken and sent to the participants.
Figure 14: Principle Investigator Dr. Robert Schlub and Pest Management students at an ironwood tree workshop held at the University of Guam on February 28, 2020.
½ Day Workshop February 29, 2020
A half-day workshop was attended by 25 participants including farmers, property owners, home gardeners, professionals, and students at the University of Guam’s College of Natural and Applied Sciences’ Agriculture and Life Science building on Saturday, February 29, 2020 from 8:30 to 11:30 am (Figure 15). The focus of the workshop was to educate the community about the ironwood tree, the ironwood tree decline (IWTD) and the current response to address the decline.
PowerPoint was used for all workshop presentations/discussions. Extension Associate DonaMila Taitano opened the workshop with a discussion of this grant’s objectives. Principle Investigator Dr. Robert Schlub then gave a presentation in regards to ironwood trees on the world stage and on Guam including: distribution, uses, economic value, and ecological value. Then Dr. Schlub discussed threats to the ironwood trees around the world and on Guam including: economic losses and ecological losses, uniqueness of ironwood trees on Guam, ironwood tree diseases, and the importance of increasing the genetic diversity of Guam’s ironwood trees. Finally, DonaMila Taitano discussed planting and care of the ironwood tree and gave out a total 30 ironwood sapling varieties from different worldwide locations for those requesting the trees.
A pre-test and a post-test were given to the participants to determine the knowledge gained. The results were significant, with an average score on the pre-test of 33% correct and an average score on the post test of 88%.
Workshop participants were asked to evaluate the workshop on a scale of 1 to 5 in different categories, with one being poor and five being excellent: 100% rated information provided at 4 or 5 (82% at 5); 100% percent rated quality of speakers at 4 or 5 (90% at 5); 82% rated overall organization at 4 or 5 (55% at 5); 100% rated overall benefit from attending the workshop at 4 0r 5 (77% at 5).
Figure 15: 15 of the 25 participants who attended the WSARE funded ironwood tree workshop on February 29, 2020.
Obj. 4 Sub 2: UOG/UH/LSU: PI and Extension/outreach representative in the third year will conduct a four-day workshop/conference on bacterial wilt and other components of IWTD.
From January 3-7, 2022, a four- and a half-day workshop/conference was held at the University of Guam.
Part one: Building Resiliency of Trees in the Northern Mariana Islands Workshop
A full-day workshop was held for 19 agricultural professionals on January 3, 2022. Participants included agricultural professionals from Guam: 4 from Guam Department of Agriculture, 6 from Guam Plant Extinction Prevention Program (GPEPP), 4 from Colorado State University Center for Environmental Management of Military Lands (CEMML), 4 from University of Guam, and 1 from Andersen Air Force Base, Guam (Figure 16). Four agricultural professionals from the Northern Mariana Islands (including forestry and extension agents from Saipan, Rota, and Tinian) were invited to the workshop, but were unable to travel due to COVID restrictions. The workshop was held at the University of Guam’s College of Natural and Applied Sciences’ Agriculture and Life Science building on Monday, January 3, 2022 from 8:00 am to 4:30 pm. The workshop was presented by Dr. Robert Schlub of the University of Guam and guest presenter Dr. Anand Persad of ACRT Services. The focus of the workshop was to update the citizens of Guam and the Northern Mariana Islands on the health of its Ironwood tree, and to provide them with tree care practices that can be used to reduce the impact of pests and diseases in Ironwood as well as other trees of the region.
The following was the agenda for the workshop: 2022 Building Resiliency Workshop
Activities included morning classroom instruction, which covered topics such as Guam’s tree species, plant health care, and soil-borne bacteria and fungi impacting the health of Guam’s trees. In the afternoon, participants were taken on a short walking tour of campus where they examined ironwood trees for symptoms of ironwood tree decline and the presence of other decline predictors/contributors (Figure 17). After the tour, participants spent the rest of the day in the lab learning various activities, including how to test samples for Ralstonia solanacearum, how to identify major termites that attack ironwood trees, examining fruiting structures of wood-rotting fungi under a microscope, examining tree disks for bacterial ooze and wetwood, and how to propagate ironwood trees.
Figure 16. Participants, presenters, and staff who attended the WSARE-funded Tree Resiliency workshop at UOG in January 2022.
Figure 17. Dr. Robert Schlub talks to participants about symptoms of ironwood tree decline as they tour the UOG Ironwood Tree Demonstration Garden.
Part two: Ironwood (Casuarina equisetifolia) decline conference
A 3.5 day in-person and virtual ironwood tree decline (IWTD) conference was held from Tuesday, January 4 to Friday, January 7, 2022 at the University of Guam Agriculture and Life Sciences building. Known ironwood tree researchers from across the globe were invited to attend in-person or virtually via zoom. The International Union of Forest Research Organizations (IUFRO) also posted an advertisement for the University of Guam IWTD conference on their twitter and Facebook pages, where an additional 300-350 people were notified of the conference and how to contact UOG if they were interested in attending virtually. Four researchers were able to attend the conference in-person: Dr. Anand Persad of ACRT Services, Dr. Sujan Paudel of the University of Hawaii, Dr. Claudia Husseneder of Louisiana State University, and graduate student Garima Setia of Louisiana State University. Many of the invited international researchers were unable to travel due to COVID restrictions. Virtual participants included researchers from the University of Hawaii, University of Florida, Louisiana State University, University of Pretoria (South Africa), the Indian Council of Forestry, the Chinese Academy of Forestry, the Commonwealth Scientific and Industrial Research Organization (Australia), and the U.S. Forest Service. During the two days of lectures, 31 people participated virtually via zoom and 4 people participated in-person, for a total of 35 participants. 17 formal presentations were given over the 3.5 day conference, including presentations by in-person and virtual attendees, and national and international researchers. The conference focused on exchange of knowledge and research discovery to ameliorate the impact of bacterial wilt in Casuarina equisetifolia, and unraveling the roles of Ralstonia solanacearum species complex, Ganoderma australe, wetwood bacteria, and termites in the decline of Guam’s ironwood.
The following was the agenda for the ironwood tree decline conference: 2022 IWTD Conference_FINAL Agenda
The first day of the conference (Tuesday, January 4th) was spent giving the in-person researchers a tour of Guam’s ironwood decline sites across the island. Sites included the Guam National Wildlife Refuge (north) and the UOG Ija Experiment Station (south) (Figure 18). Participants were able to see the effects of IWTD in-person, ask questions, and take photos.
The second day of the conference consisted of in-person and virtual classroom lectures. In-person participants were able to connect with virtual participants via zoom (Figure 19, Figure 20). Lecture topics and presenters can be seen within the agenda above.
The third day of the conference consisted of in-person and virtual lectures in the morning (Figure 21). In the afternoon, in-person participants departed for a field trip to Bernard Watson’s farm. At the farm, participants were able to see first-hand the effects of IWTD on farm windrows (Figure 22). Participants also saw new windrow plantings (Figure 23). Lastly, an ironwood tree which was a known positive of R. solanacearum was cut down at the trunk and participants were able to see evidence of wetwood, and bacterial ooze (R. solanacearum) contributing to IWTD (Figure 24).
The fourth (and final) day, WSARE Research Project team members who were able to attend in-person held an informal discussion of project goals and accomplishments.
Figure 18. In-person conference attendees pose for a picture with Dr. Robert Schlub on Ritidian beach, Guam National Wildlife Refuge.
Figure 19. Sujan Paudel of the University of Hawaii gives an in-person presentation on Genetic diversity and genealogy of the Ralstonia solanacearum species complex associated with Ironwood decline in Guam. Virtual participants could view his presentation via zoom meetings.
Figure 20. Indian Council of Forestry researcher Dr. Abel Nicodemus gives a virtual presentation via zoom. Dr. Nicodemus and his colleague Dr. Arumugam Karthikeyan are visible in the upper right-hand corner of the screen.
Figure 21. Dr. Anand Persad gives a presentation on “Climate and Island States.”
Figure 22. An ironwood tree windrow seen at Bernard Watson’s farm. This windrow has not yet been fully decimated by IWTD.
Figure 23. Researchers examine planted ironwood tree windrows at Bernard Watson’s farm. These trees are approximately 3 years old.
Figure 24. Extension agents show researchers the wetwood and signs of bacterial ooze on a freshly cut-down ironwood tree stump. This tree was known positive for R. solanacearum.
Obj 1 Yr 1-2: UOG Ed: Through tree plantings, educate the public on the importance of planting new off-island cultivars (seedlots from international provenance trails)
Results: Obj. 1, Sub-obj. 1, UOG: Plant trees of mixed cultivars in 10 visually deteriorating windrows across Guam.
Property owners were educated on how to refurbish windrows. Ten windrows were refurbished with 6-15 trees each, for a total of 138 trees planted.
Results: Obj. 1, Sub-obj. 2, UOG: Construct 15 new ironwood agroforestry projects using 150 trees consisting of a mixture of tree cultivars.
Property owners were educated on the best tree care and management practices during planting. Fifteen new windrows were created consisting of 10 trees each, for a total of 150 trees.
Obj. 4 Yr 2-3: UOG Ed: Restoring ironwood as an agroforestry species in Guam through awareness and action of the local and scientific communities.
Results: Obj. 4, Sub-obj. 1, UOG: Through two ½ day workshops, attendees will learn about the care of the ironwood trees and its uses in agroforestry. Ironwood trees will be given away.
1st ½ day workshop:
Eleven workshop participants successfully planted ironwood seeds into cone planters and ironwood seedlings into soil.
The participants also filled out their own ironwood tree evaluation spreadsheet. Information they collected and recorded included decline severity (DS= 0-4), location, altitude, GPS, height, circumference at breast height (CBH), termite activity, presence or absence of Ganoderma conk, and tested for the presence or absence of Ralstonia using Agdia Rs immunostrip test kits.
Lastly, participants inoculated tomato seedlings with Ralstonia isolates collected previously. Half of the plants inoculated with Rs developed severe bacterial wilt symptoms, while no symptoms developed on the plants that weren’t inoculated.
2nd ½ day workshop:
Twenty-five workshop participants took a pre-test and a post-test covering workshop topics to determine the knowledge gained. The results were significant, with an average score on the pre-test of 33% correct and an average score on the post test of 88%. Topics included: ironwood trees on the world stage and on Guam including: distribution, uses, economic value, and ecological value; threats to the ironwood trees around the world and on Guam including: economic losses and ecological losses; uniqueness of ironwood trees on Guam; ironwood tree diseases; and the importance of increasing the genetic diversity of Guam’s ironwood trees.
Results: Obj. 4 Sub 2: UOG/UH/LSU: PI and Extension/outreach representative in the third year will conduct a four-day workshop/conference on bacterial wilt and other components of IWTD.
Part one: Building Resiliency of Trees in the Northern Mariana Islands Workshop
Nineteen agricultural professionals from Guam and the Northern Mariana Islands attended a tree resiliency workshop. The attendees were updated on the current health of Guam’s Ironwood trees, and educated about tree care practices that can be used to reduce the impact of pests and diseases in Ironwood as well as other trees of the region.
Part two: Ironwood (Casuarina equisetifolia) decline conference
The 3.5 day IWTD conference was a success. The conference focused on exchange of knowledge and research discovery into the impact of bacterial wilt in Casuarina equisetifolia, and unraveling the roles of Ralstonia solanacearum species complex, Ganoderma australe, wetwood bacteria, and termites in the decline of Guam’s ironwood. Thirty-five national and international researchers participated in the conference and 17 formal presentations were given over the 3.5 days (Table 6).
Table 6. Formal presentations given by national and international ironwood tree researchers at the 2022 Guam IWTD Conference.
|
Presentation Title |
Presenter |
Organization |
|
“Overview: Guam’s Ironwood (C. equisetifolia) and its decline” |
Dr. Robert Schlub |
University of Guam
|
|
“Statistical applications into Ironwood trees (C. equisetifolia) decline on Guam and varietal selection” |
Karl Schlub
|
Louisiana State University (Thesis) |
|
“Overview of Ralstonia solanacearum species complex (RSSC) and population biology” |
Dr. Mohammad Arif
|
University of Hawaii
|
|
“Phylogenetic characterization and genealogy of strains in Ralstonia solanacearum species complex (RSSC) associated with ironwood decline in Guam” |
Sujan Paudel
|
University of Hawaii (Thesis) |
|
“Genome evolution of Ralstonia solanacearum species complex (RSSC) associated with ironwood decline in Guam” |
Dario Arizala
|
University of Hawaii (Thesis) |
|
“Field deployable recombinase polymerase amplification assay and other methods used at the University of Hawaii to differentiate Ralstonia solanacearum species complex and related bacteria from Ironwood samples” |
Dr. Shefali Dobhal
|
University of Hawaii (Thesis) |
|
“Comparisons between Ironwood tree decline and bacterial wilt in China and India” |
Dr. Robert Schlub |
University of Guam
|
|
"Occurrence and control of bacterial wilt in C. equisetifolia coastal shelterbelts in Southern China" |
Dr. Zhang Yong |
Chinese Academy of Forestry
|
|
“Casuarina research in China” |
Dr. Zhong Chonglu |
Chinese Academy of Forestry (Retired) |
|
“Disease management in high density clonal plantations of Casuarina in India” |
Dr. Abel Nicodemus |
Indian Council of Forestry Res. & Ed. |
|
“Possible biocontrol agents for bacterial wilt disease in Casuarina” |
Dr. Arumugam Karthikeyan |
Indian Council of Forestry Res. & Ed. |
|
Bacterial wilt of Eucalyptus |
Dr. Teresa Coutinho |
University of Pretoria |
|
“Bacterial wilt in Florida and diagnostic procedures used at the University of Florida” |
Dr. Carrie Harmon |
University of Florida |
|
“Introduction into termite biology, feeding and symbiosis and termites of Guam’s Ironwood trees (C. equisetifolia)” |
Dr. Claudia Husseneder |
Louisiana State University |
|
“Investigation of potential role of termites as pathogen vectors in the decline of Ironwood trees (C. equisetifolia) in Guam” |
Garima Setia |
Louisiana State University (thesis) |
|
“Tree varieties in place of dying Casuarina” |
Anand Persad |
ACRT Services |
|
“Climate and Island States” |
Anand Persad |
ACRT Services |
Information about the IWTD Conference and how to receive a link to join the virtual presentations via zoom were posted to the International Union of Forest Researchers Organizations (IUFRO) Twitter, Facebook, and Linkedin social media pages. Although there were no requests for the zoom link, the posts were seen by at least 300-350 people.
Education and Outreach Outcomes
This project arose out of the concern that the death of ironwood trees across Guam would result in the abandonment of the longtime sustainable agriculture practice of planting ironwood trees for windbreaks, reforestation, and erosion protection programs. As a result of research discoveries, this project has developed stewardship practices that can be implemented to sustain the longtime viability of the ironwood tree, a natural resource important to the US and the Pacific region.
As a result of several education and outreach activities, we have found that demonstration plots and workshops with hands-on activities are excellent ways to affect stakeholders’ understanding of agricultural sustainability. Whereas, dissemination of agricultural research results were best achieved through a conference of researchers. Activities included two ½ day workshops held on February 28 and 29, 2020, and a four-and-a-half-day workshop/conference held on January 3-7, 2022. The February 28 workshop was for UOG Pest Management students, and the February 29, 2020 workshop was for farmers, property owners, home gardeners, professionals, and students of the University of Guam College of Natural and Applied Sciences.
Participants of the February 28 workshop were exposed to a variety of practices and techniques used in studying ironwood tree decline. The focus of the February 29 workshop was to educate the community about the ironwood tree, ironwood tree decline (IWTD), and the current response to address the decline. Based on the assessment of knowledge gained by participants in the February 29 workshop, we have found that the project has affected stakeholders' understanding of the importance of healthy stands of ironwood trees in agricultural sustainability of Guam and the islands of the region.
The four-and-a-half-day workshop/conference was divided into two parts. On January 3, 2022, a full-day workshop was conducted on Building Resiliency of Trees in the Northern Mariana Islands. This was attended by 19 agricultural professionals. The focus of the workshop was to update the citizens of Guam and the Northern Mariana Islands on the health of their ironwood trees and to provide them with information on tree care practices that can be used to reduce the impact of pests and diseases on ironwood as well as other trees of the region. The second portion was a 3.5-day ironwood tree decline (IWTD) conference. It was held from Tuesday, January 4 to Friday, January 7, 2022. The workshop was held in person and virtually. The conference focused on the exchange of knowledge and research discovery to ameliorate the impact of bacterial wilt in ironwood and unraveling the roles of Ralstonia solanacearum species complex, Ganoderma australe, wetwood bacteria, and termites in the decline of Guam’s ironwood.
As a result of this project, recommendations for education and outreach to reduce ironwood bacterial wilt and its associated disease, ironwood tree decline, are as follows:
- Whenever possible, trees from C. equisetifolia seed lots from the Commonwealth Scientific and Industrial Research Organization (CSIRO) Australian Tree Seed Center (ATSC) should be planted. As a result of cross-pollination, the gene pool of Guam’s trees will be strengthened.
- When using a lawnmower or weed trimmer, care should be taken not to wound tree roots or the base of trees. Wounds create entryways for infection by plant pathogens from the environment or from contaminated lawn equipment.
- To reduce transmission of Ralstonia and wetwood bacterial ooze when pruning, all tools should be disinfected.
- Trees in severe decline should be removed as a means to protect nearby healthy trees.
- Planting healthy young trees is the quickest way to replace trees in decline. Any young, healthy ironwood tree will do; it can be from seed of a Guam tree or one of the cultivars from ATSC. The tree can be planted near a declining tree, provided the distance is great enough to prevent any comingling of their roots; 100 ft should be sufficient. The new tree can be planted in the same spot where a tree died, provided there are no tree remnants present such as pieces of trunk, branches, or roots.
- Agroforestry
Ironwood tree genetic diversity and bacterial disease