2016 Annual Report for GNE16-130
Uncovering the genetic control of cold resistance and adapting winter survival models for Northeast grape production
Summary
Cold hardiness studies are of great importance for the Northeastern US, where winter weather can lead to damage of perennial crops. Grapevines can suffer from cold injury in several ways, of which one of the most common is the injury to fruiting buds. The purpose of the current study is to identify genes responsible for cold hardiness aspects, and to adapt models for prediction of cold hardiness in grapevine buds during the winter. To evaluate this, buds are being collected from vineyards in the Finger Lakes Wine Region in NY. The buds are being used in differential thermal analysis (DTA), yielding a measure of the cold hardiness at given times. Buds are also being collected for RNA extraction. To date, there have been 13 weekly collections. Buds for water content have not been dried, and no RNA extractions have been performed, however all DTA data has been collected and compared to a prediction model based on previous years’ data.
Objectives/Performance Targets
The proposal contained two main objectives:
- Generate or adapt a prediction model for cold hardiness for the Northeastern United States, using DTA data from the Finger Lakes wine region in New York.
- Evaluate gene expression changes throughout the winter as a response to temperature shifts to identify specific genes that may serve as markers for cold hardiness aspects and enriched pathways related to cold hardiness.
From objective 1, DTA data have been collected for 13 weeks to date for V. vinifera ‘Cabernet Franc’ and ‘Riesling’, and V. interspecific hybrids ‘Cayuga White’ and ‘Marquette’, from Anthony Road Wine Co. V. vinifera ‘Cabernet Sauvignon’ was added later, and has been collected for 10 weeks from Ravines Wine Cellars. The DTA data have been compiled and organized with weather data for future analysis. Buds collected for water content will be freeze dried once all collections have ceased. Additionally, single node cuttings were collected from ‘Cabernet Franc’, ‘Cabernet Sauvignon’, ‘Riesling’, and ‘Sauvignon Blanc’, as well as 2 genotypes of V. riparia and one of V. aestivalis for controlled acclimation in growth chambers. The temperatures used were slightly different than those proposed to match key temperatures in chill accumulation models: 2, 4, 7, 11, and 22 °C, and complete dark was used instead of a 12 h photoperiod. DTA data have been collected on these for 10 weeks.
On objective 2, buds have been collected for 13 weeks from ‘Riesling’ for RNA extraction. Once the dormant season is finished and all samples have been collected, as described in the proposal timetable, RNA extractions will be performed and libraries prepared for RNA sequencing.
Accomplishments/Milestones
To this date, about half of the field collections have been performed. From these, the DTA data will be used to develop a model for prediction of cold hardiness. Using last year’s data, we have been comparing the predicted values with the measure and have identified that the model requires further adaptation. The high-density data that is being collected will be crucial in developing a model with high confidence for the region that will likely be adaptable to other places.
For the controlled acclimation experiment, the DTA data has shown no shift in cold hardiness regardless of the temperature exposure and cultivar. This goes against currently available predictive model and indicates that other factors are playing a role. Because constant temperatures were not affecting cold hardiness, we decided to change the temperature profile in one of the chambers to have temperature oscillations, such as in the field, to study whether that played a role. The new cold chamber had a temperature profile of 1.5 – 7 – 12.5 – 7 °C for periods of 6 h in each temperature. With this, the average temperature was still 7 °C. Material from the constant 2, 7, and 11 °C were moved into the new oscillating temperatures chamber. After 4 weeks, it appears that the temperature oscillation leads to further acclimation of at least 4 of the 7 genotypes tested. These results have a great impact on the development of the prediction model, since it may have elucidated the reason why current models do not yield good results in the NE: they use average daily temperatures and therefore neglect the effect of daily temperature cycles.
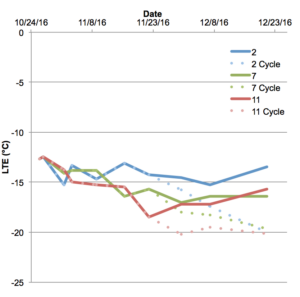 Figure 1. Low temperature exotherm (LTE) data obtained through differential thermal analysis of V. aestivalis exposed to 2, 7, 11 °C followed or not by cycling 1.5 – 7 – 12.5 – 7 °C in 24 h periods demonstrates the increase in cold hardiness after exposure to cycling temperatures.
Figure 1. Low temperature exotherm (LTE) data obtained through differential thermal analysis of V. aestivalis exposed to 2, 7, 11 °C followed or not by cycling 1.5 – 7 – 12.5 – 7 °C in 24 h periods demonstrates the increase in cold hardiness after exposure to cycling temperatures.
Impacts and Contributions/Outcomes
Because this project was only initiated in October, there have been no impacts on current practices. We expect, however, that this project will yield a useful model to predict cold hardiness based on the weather data. With the model developed, farmers can be aware of damage occurring and plan for pruning accordingly in order to reduce the impacts of the cold damage in the yields. If methods are developed in the future for increasing cold hardiness through cultural practices, this model will help in the decision-making. Additionally, if genes associated with cold hardiness are found, these can be used as markers in the future to develop new cultivars for regions where there are difficulties associated with winter weather. Although major impacts have not yet occurred, we have found that temperature oscillations might be playing an important role in the development of the hardiness phenotype observed, which will be important in the planning of future controlled acclimation experiments, as well as in the development of the prediction model.
Collaborators:
Research Geneticist
USDA-ARS Grape Genetics Research Unit
630 W. North St.
Geneva, NY 14456
Office Phone: (315) 787-2463