Final report for GNC19-278
Project Information
Fire blight, caused by the bacterium Erwinia amylovora, is a devastating bacterial disease of apples in Ohio (OH). Fire blight is a systemic disease and one of the most difficult diseases of apple to manage. Current management strategies to minimize production losses include cultural practices and the use of antibiotics. Antibiotics, including streptomycin, oxytetracycline, and kasugamycin, are highly effective in reducing E. amylovora populations during bloom, but continuous use of them is not sustainable. For example, resistance to the most effective of the three antibiotics-streptomycin, has been reported in many orchards in the northeastern United States (US). In addition, the use of antibiotics in agricultural has raised legitimate concerns about the future effectiveness of antibiotics to control human diseases through the transfer of antimicrobial resistance genes to human pathogens.
Biological plant disease control can be integrated into fire blight management programs; reducing or eliminating the reliance on antibiotics. Previous studies conducted in northeastern US states have demonstrated that pairing species of Bacillus with antibiotics can minimize fire blight incidence. Currently, the use of biological control agents (BCAs) to manage fire blight is mostly limited to organic producers and backyard gardeners. Commercial and conventional apple producers have shied away from BCAs for fire blight control because of the lack of consistent efficacy data, the number of applications required to achieve a high level of efficacy, and their short shelf life. Despite these challenges, there continues to be an interest in BCAs for disease management, which probably reflects continued consumer concerns about pesticide use and environmental and human health. However, the identification and implementation of microorganisms with biocontrol activity is slow and requires extensive laboratory and field testing before BCAs make it to the market. In this study, we will evaluate the suppression of E. amylovora isolates with newly identified and novel antagonistic microorganisms. The proposed study aims to characterize and evaluate the efficacy of these novel BCAs for biological control of fire blight in Ohio with the long-term goal of providing sustainable alternatives to antibiotics. Laboratory and field studies to evaluate the efficacy of novel BCAs will be conducted. Field days and grower meetings and conferences, will facilitate apple farmers involvement in the learning process and understanding of the use of BCAs as a sustainable alternative to antibiotics.
Outcomes of this project will directly benefit apple growers in OH who have continuous losses due to fire blight disease and use antibiotics as the main strategy to combat the disease.
Learning outcomes: We expect to: 1) identify effective BCAs programs to reduce fire blight incidence and spread in the orchard and 2) increase grower awareness of integrating BCAs into a fire blight management program.
Action outcomes: Apple growers will 1) adopt an effective management approach that can include the use of biocontrol agents to manage fire blight and; 2) reduce their dependency on antibiotics (e.g. streptomycin) use for fire blight management.
Research
Materials and Methods
1. Bacterial strains and growth inhibition assays. An in vitro assay was conducted to test the ability of 50 Pseudomonas spp. (C. Taylor, The Ohio State University, Dept. Plant Pathology; Table 1), previously isolated from plant, water and soil samples (Tao et al. 2015) in inhibiting the growth of E. amylovora. Four E. amylovora strains (MLI90-17; MLI181-18; MLI200-18; MLI217-18) were isolated from symptomatic shoot tissue and were sensitive to streptomycin (Jimenez et al. 2020). Zone of growth inhibition assays were conducted according to Kirby et al. 1956 and Bauer et al. 1966. Pseudomonas spp. and E. amylovora were recovered from glycerol stocks stored at -80°C and plated onto Luria-Bertani (LB) and nutrient broth yeast (NBY) agar media, respectively and incubated at 28°C for 48 hrs. E. amylovora and Pseudomonads spp. inoculum was prepared for each strain by suspending 250 µl in 250 ml of sterile LB at 0.7% and incubate the suspension for 24 hrs at 28°C. Seven ml of each E. amylovora inoculum (~108 CFU/ml; OD600 1.0-1.2) was then evenly distributed on a petri plate (VWR International Corp. Batavia, IL) and two sterile 6mm filter paper disks (Fisher Scientific, Waltham, MA) were placed on top of the LB agar plate. Each filter paper disk received 20 µl of one strain of Pseudomonas spp. adjusted to an OD600 ~1.0. The process was repeated until all 50 strains of Pseudomonas spp. were tested. LB agar without E. amylovora served as the negative E. amylovora control and sterile water served as the Pseudomonas spp. negative control. Kanamycin (50 mg/ml) was applied to the filter disks as a positive control. The plates were incubated at 28°C for 24 to 48 hrs and the zones of growth inhibition were measured (mm) using a graduated ruler (Universal, Atlanta, GA) by measuring the diameter. The experiment was repeated twice, and each experiment was considered a replicate. Statistical differences in the size of the inhibition zones between each E. amylovora strain and each Pseudomonas spp. strain were determined using PROC GLM procedure in SAS (SAS version 9.4) and means were separated by Fisher's Protected LSD (LSD α=0.05).
Table 1. List of Pseudomonas species including group number and the source from which the strains were isolated from, used in this study.
|
Speciesa (Groupb) |
Strain Identification |
Source |
Reference |
|
P. vranovensis (II) |
15D11 |
Water |
Mavrodi et al. 2012 |
|
P. chlororaphis (IV) |
14D6 48G9 14B11 48B8 |
Water Soil Water Soil
|
Tao et al. 2015 Tao et al. 2015 Mavrodi et al. 2012 Tao et al. 2015 |
|
P. protogens (V) |
14B2 12H11 15G2 15H3 38G2 1C5 1B1 Darke 15H10 15G6 Wayne Clinton 1F2 |
Water Water Water Water Soil Water Water Soil Water Water Corn Soil Water |
Tao et al. 2015 Tao et al. 2015 Mavrodi et al. 2012 Mavrodi et al. 2012 Mavrodi et al. 2012 Tao et al. 2015 Mavrodi et al. 2012; Tao et al. 2015 Tao et al. 2015 Tao et al. 2015 Mavrodi et al. 2012 Mavrodi et al. 2012 Tao et al. 2015 Tao et al. 2015 |
|
P. poae (VI) |
29G9 36C8 |
Herbarium Soil |
Mavrodi et al. 2012 Tao et al. 2015 |
|
P. rhodesiae (VI) |
88A6 90F12-1 |
Soil Soil |
Mavrodi et al. 2012 Mavrodi et al. 2012 |
|
P. brassicacearum (VII) |
Wood3 36B7 36D4 93G8 Wood1 93D8 |
Soybean Soil Soil Soybean
Soil |
Mavrodi et al. 2012 Tao et al. 2015 Mavrodi et al. 2012 Tao et al. 2015
Tao et al. 2015 |
|
P. brassicacearum (VIII) |
37D10 48H11 |
Soil Soil |
Tao et al. 2015 Mavrodi et al. 2012 |
|
P. brassicacearum (IX) |
38D7 38D4 |
Soil Soil |
Tao et al. 2015 Tao et al. 2015 |
|
P. frederiksbergensis (IX) |
94G2 36C6 39A2 37A10 38F7 37A11 |
Soil Soil Soil Soil Soil Soil |
Tao et al. 2015 Tao et al. 2015 Mavrodi et al. 2012 Mavrodi et al. 2012 Tao et al. 2015 Tao et al. 2015
|
|
P. lini (IX) |
48C10 |
Soil |
Mavrodi et al. 2012 |
|
P. fluorescens (X) |
36F3 48D1 28B5 24D3 2F9 36G2 36B3 89F1 90D7A 90F12-2 48D5 |
Soil Soil Herbarium Herbarium Water Soil Soil Soil Soil Soil Soil |
Tao et al. 2015 Mavrodi et al. 2012 Mavrodi et al. 2012 Tao et al. 2015 Tao et al. 2015 Tao et al. 2015 Tao et al. 2015 Tao et al. 2015 Tao et al. 2015 Mavrodi et al. 2012 Mavrodi et al. 2012 |
a All Pseudomonas species were provided by C. Taylor, The Ohio State University, Dept. Plant Pathology.b Group number refers to phylogenetic cluster sequence analysis of ten housekeeping genes, previously described by Tao et. al 2015.
2. Colonization of apple blossoms by Pseudomonas spp. Two Pseudomonads strains (P. protogens strain 1B1 and P. brassicacearum strain 93G8) with high inhibitory activity in vitro against all four strains of E. amylovora (MLI90-17; MLI181-18; MLI200-18; MLI217-18) were selected to evaluate the colonization of apple blossoms. Spontaneous rifampicin resistant mutants of 1B1 and 93G8 were selected using a laboratory standard gradient plate technique (Gerhardt et al. 1994). Briefly, 15 ml of LB agar was dispensed into a sterile 100 mm Petri dish (100 mm) that had one edge slightly elevated. Once the agar cooled, the plates were leveled and 15 ml of LB supplemented with 100 µg/ml of rifampicin (TCI America, Portland, OR), dispensed over the cooled LB agar to generate a concentration gradient of rifampicin from 0 to 100 µg/ml. 1B1 and 93G8 were grown for 24 hrs in LB broth at 28°C with shaking (~165 rpm) and then 0.1 ml of the bacterial suspensions (~108 cfu/ml) were spread onto the gradient plates and incubated for 36-48 hrs at 28°C. Four individual colonies of each strain growing on the region of the gradient with a high concentration of rifampicin were selected and streaked onto new gradient plates. After 36-48 hrs at 28°C four new colonies were again selected from the region of the gradient with a high concentration of rifampicin and stored at -80°C in 30% glycerol and LB broth supplemented with rifampicin (30 µg/ml) (1:1 V:V). The species identification of the selected mutants (1B1rif and 93G8rif) were confirmed using strain-specific PCR with the primers PLTC1 (5’-AACAGATCGCCCCGGTACAGAACG-3’) and PLTC2 (5’-AGGCCCGGACACTCAAGAAACTCG-3’) for 1B1 (de Souza and Raaijmakers 2003) and HFG (5’-GCACTTCGATGTGGTCATCA-3’) and HFH (5’-GATGGGTTTGAACAGGCAAT-3’) for strain 93G8 (Tao, unpublished). PCR assays were performed in 25 µl reaction volumes and consisted of 12.5 µl of Green GoTaq Master Mix (Promega Corp., Madison, WI), 9.5 ml of nuclease free H2O, 1 µl each of forward and reverse primers and 1 µl of whole cell extracted DNA. Cycling parameters were 3 min at 94°C; followed by 36 cycles of 94°C for 30 s, annealing at 65°C for 30 s (for 1B1) and 55°C ( for 93G8), and 72°C for 60 s; followed by a final extension at 72°C for 5 min. PCR products were separated using gel electrophoresis with 1.5% agarose gels in 1× TBE buffer (44.5 mM Tris-acetate and 1 mM EDTA, pH 8.0) at 90 V for 90 min. DNA stained with GelRed (Biotium Inc. Fremont, CA) was visualized under Axygen® Gel Documentation System (Corning Incorporated Life Sciences, Tewksbury, MA). Wildtype Pseudomonas 1B1 and 93G8 strains served as the positive controls and nuclease free water served as negative control.
Four (n=4), 4-year old ‘Golden Delicious’ (susceptible to fire blight) on Bud 9 rootstock (Wafler Nursery, Wolcott, NY) produced in 25-gallon pots containing sterilized Wooster silt loam were placed in dark cold storage (4 °C) for 75 days to achieve a minimum of 1800 chilling hrs. The trees were then transferred to a greenhouse on 1 Feb 2021 and forced into full bloom using the following environmental conditions for 7 days: 24°C during the day and 18°C at night with supplemental light for a photoperiod of 16 hrs (6 am to 10 pm). After 7 days, the day and night temperature were set at a constant 18°C. Trees reached silver tip by 3 Feb 2021, green tip by 6 Feb 2021, half-inch green by 9 Feb 2021, pink by 15 Feb 2021, and by 18 Feb 2021 the trees were at least 80% bloom (Figure 1). The average relative humidity in the greenhouse from dormant stage to bloom was 16.3% and the average temperature was 21.7 °C. Inoculum of 1B1rif or 93G8rif was prepared by growing the strains on LB agar amended with rifampicin (50 mg/ml) for 48-72 hrs at 28C and then suspending the bacteria in sterile water and adjusting the OD600 to 1.0 (~108 CFU/ml). Once the flowers reached ~80% bloom they were inoculated. Across the four apple trees 12 flower clusters per strain were randomly selected and sprayed with 1mL of inoculum using a Preval sprayer (Chicago Aerosol, Coal City, IL). To minimize cross-contamination between the strains and the non-treated controls (water only) each apple cluster was protected during spraying by surrounding the cluster with a disposable 0.6 mm paper plate. Flower clusters were hand-picked at 0, 1- and 3-days post inoculation, placed and sealed in 59 ml Whirl-Pak bags (Nasco, Fort Atkinson, WI) and transferred to the lab for processing. The total flower cluster weight in grams and weight without leaves were recorded for each cluster and then 8 mL of 1X PBS (pH 7.0) buffer was added to each sample and the samples were pulsified for 2 min using a Microbiology International Pul100 Pulsifier (Microgen Bioproducts Ltd, UK). The supernatant was then transferred to 15 mL sterile plastic centrifuge tubes and centrifuged at 4, 122 x g for 15 min. The supernatant was discarded, and the pellet was re-suspended in 1 mL of 1X PBS and 10-fold serially diluted to 10-6. Five ul of each dilution was spot plated in duplicate on LB amended with rifampicin (50 µg/ml) and cycloheximide (100 µg/ml). Colonies within each droplet were counted after 24 and 48 hrs of incubation at 28 °C. The CFU/g flower tissue was calculated, and the data were log transformed. Statistical differences in the number of CFU/g flower tissue between the treated and non-treated flowers were determined using PROC GLM procedure in SAS (SAS version 9.4) and means were separated by Fisher's Protected LSD (LSD α=0.05). The percent increase or decreased in CFU/g tissue at 3- and 7-days post-inoculation, compared to 0-days post-inoculation, was also calculated. The entire experiment was conducted twice but only the results of the first experiment are presented in this dissertation.
Figure 1. Phenological stages of cv. Golden Delicious. A Silver tip. B Green tip. C Half-inch green. D Tight cluster. E Pink. F Bloom
3. Efficacy of Pseudomonas spp. in controlling blossom blight infections Two Pseudomonad strains (P. protogens strain 1B1 and P. brassicacearum strain 93G8) with high inhibitory activity in vitro against E. amylovora were evaluated for their ability to inhibit blossom blight infections in planta. Flower clusters on cv. Golden Delicious were treated 1B1, 93G8, or streptomycin when at least ~80% of the blossoms were open. After 72 hours the flowers were inoculated with E. amylovora Ea110 (OD600 to 1.0 ;~108 CFU/ml). Non-treated, non-inoculated control clusters ( n=6) were included in the experiment. The experiment was conducted in March and the trees were maintained in a greenhouse with no supplemental lighting or temperature control. Flower clusters were monitored daily for 30 days. The number of clusters with fire blight symptoms were counted and percent disease incidence calculated. The clusters were removed by cutting approximately 2.5 cm below the cluster (Figure 2) and ~ 1 cm between the terminal cut and cluster was removed, weighed and isolations of E. amylovora on the selective medium CM were conducted and the CFU/g tissue calculated. Data were log transformed and statistical differences in the number of CFU/g tissue between the treated clusters were determined using PROC GLM procedure in SAS (SAS version 9.4) and means were separated by Fisher's Protected LSD (LSD α=0.05). The experiment was not repeated.

Results
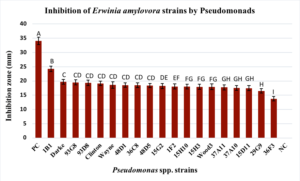
1. Inhibition of Erwinia amylovora in vitro. Among the 50 strains of Pseudomonas spp. screened for inhibitory activity 31 did not have a zone of inhibition against any of the four strains of E. amylovora (Table 2). None of the Group IV (P. chlororaphis), P. rhodesiae Group VI, P. brassicacearum Group VIII or IX strains, or the one Group IX (P. lini) strain had a zone of inhibition against the E. amylovora strains and 73% of strains from Group X (P. fluorescens) also did not have any inhibitory activity against E. amylovora. For the remaining 19 strains of Pseudomonas spp., zones of inhibition against E. amylovora strains MLI90-17, MLI181-18, MLI200-18, and MLI217-18 ranged from 27-13 mm, 25-14 mm, 25-17 mm and 24-16 mm, respectively (Table 2). All but five of the Group V P. protegens strains (14B2, 12H11, 38G2, 1C5, and 15G6) resulted in a zone of inhibition, including P. protegens strain 1B1, which had on average the largest zone of inhibition for all four E. amylovora strains (MLI90-17= 27 mm, MLI181-18= 25 mm, MLI200-18= 25 mm, and MLI217-18= 24 mm) (p<.0001 for all four strains).
Table 2. Mean zone of inhibition in millimeters of Pseudomonas spp. strains against four strains of E. amylovora
|
|
|
|
Erwinia amylovora strains |
|
|
|||
|
Pseudomonas spp. strain name |
Species |
Group |
MLI90-17 |
MLI181-18 |
MLI217-18 |
MLI200-18 |
Meanc (mm) |
Mean Separationc |
|
Kanamycin controla |
- |
- |
35.0 |
35.0 |
33.0 |
37.0 |
34.05 |
A |
|
1B1 |
P. protegens |
V |
27.0 |
25.0 |
24.0 |
25.0 |
24.22 |
B |
|
Darke |
P. protegens |
V |
23.0 |
20.0 |
19.0 |
21.0 |
19.66 |
C |
|
Wayne |
P. protegens |
V |
23.0 |
20.0 |
19.0 |
20.0 |
18.61 |
CD |
|
36C8 |
P. Poea |
VI |
22.0 |
19.0 |
19.0 |
19.0 |
18.50 |
CD |
|
93G8 |
P. brassicacearum |
VII |
22.0 |
20.0 |
20.0 |
20.0 |
19.50 |
CD |
|
Clinton |
P. protegens |
V |
22.0 |
19.0 |
19.0 |
20.0 |
19.11 |
CD |
|
93D8 |
P. brassicacearum |
VII |
22.0 |
21.0 |
19.0 |
20.0 |
19.33 |
CD |
|
Wood3 |
P. brassicacearum |
VII |
21.0 |
19.0 |
17.0 |
20.0 |
17.94 |
FG |
|
15H10 |
P. protegens |
V |
20.0 |
19.0 |
18.0 |
18.0 |
18.00 |
FG |
|
48D5 |
P. fluorescens |
X |
20.0 |
19.0 |
18.0 |
20.0 |
18.38 |
CD |
|
1F2 |
P. protegens |
V |
20.0 |
19.0 |
17.0 |
19.0 |
18.05 |
EF |
|
15H3 |
P. protegens |
V |
19.0 |
19.0 |
18.0 |
19.0 |
17.94 |
FG |
|
15D11 |
P. vranovensis |
II |
19.0 |
18.0 |
17.0 |
18.0 |
17.44 |
GH |
|
15G2 |
P. protegens |
V |
19.0 |
20.0 |
19.0 |
21.0 |
18.22 |
DE |
|
37A10 |
P. frederiksbergensis |
IX |
19.0 |
19.0 |
16.0 |
19.0 |
17.58 |
GH |
|
37A11 |
P. frederiksbergensis |
IX |
19.0 |
19.0 |
17.0 |
19.0 |
17.76 |
GH |
|
48D1 |
P. fluorescens |
X |
19.0 |
18.0 |
20.0 |
19.0 |
18.55 |
CD |
|
29G9 |
P. Poea |
VI |
19.0 |
17.0 |
17.0 |
18.0 |
16.50 |
H |
|
36F3 |
P. fluorescens |
X |
13.0 |
14.0 |
16.0 |
17.0 |
13.72 |
I |
|
Negative controlb |
- |
- |
0.0 |
0.0 |
0.0 |
0.0 |
0.00 |
- |
|
P-valued |
|
|
<.0001 |
<.0001 |
<.0001 |
<.0001 |
<.0001 |
|
a Kanamycin applied at 50 mg/ml.b Absence of a Pseudomonas strain (LB medium only).c Mean size of the zone of inhibition (mm) across all four strains of E. amylovora. Means followed by the same letter are not significantly different by Fisher's Protected LSD (p<0.05).d Significant differences in the size of the zone of inhibition (mm) among the Pseudomonas spp. strains were observed at p<0.05.
The two Group VI Pseudomonas poea strains 36C8 and 29G9 resulted in mean zones of inhibition, of 16.50 and 18.50 mm, respectively. The mean zone of inhibition for three strains of the Group VII P. brassicacearum Wood3, 93G8 and 93D8, against the four E. amylovora strains were 17.94 mm, 19.50 and 19.33, respectively. Only two of the six strains from Group IX, P. frederiksbergensis showed zone of inhibition, with a mean of 17.58 mm (strain 37A10) and 17.76 mm (strain 37A11). Finally, three strains of Group X P. fluorescens had zones of inhibition, one of which, strain 39F3, had the lowest mean zone of inhibition (13.72 mm) (p<.0001). Group X P. fluorescens 48D1 and 48D5 resulted in zones of inhibition of 18.55 mm and 18.38 mm, respectively. Across the four E. amylovora strains the mean size of inhibition zone for kanamycin, which served as a positive control, was 34.1 mm and was the largest compared to all of the Pseudomonads screened (Figure 2).
Figure 3. Inhibitory activity of 19 out of 50 Pseudomonads screened against E. amylovora strains MLI90-17, MLI181-18, MLI200-18, and MLI217-18 from Ohio. Bars represent the standard error of mean inhibition zones (mm) across all four strains of E. amylovora. Means followed by the same letter are not significantly different at p<0.05). PC indicates positive control (kanamycin at 50 mg/ml). NC is the negative control (no E. amylovora).
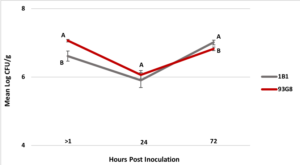
2. Colonization of apple blossoms by Pseudomonas spp. Two rifampicin resistant strains were successfully selected for up to a MIC of 50 µg/ml and PCR with the strain-specific primers confirmed that the selected strains were 1B1 and 93G8 (data not shown). Between 1 to 3 hrs (> 1 hr time point) after the bacteria were applied to the flowers the mean Log CFU/g flower tissue was 6.6 for P. protegens 1B1 and 7.0 for P. brassicacearum 93G8 (Figure 3), representing a 1.4 and 1 Log reduction in the recovery rate of each strain, respectively. After 24 hrs the mean Log CFU/g flower tissue was 5.9 for P. protegens 1B1 and 6.1 for P. brassicacearum 93G8. Finally, 72 hrs post-treatment the mean Log CFU/g flower tissue was 7.0 for P. protegens 1B1 and 6.8 for P. brassicacearum 93G8. No statistical difference (p=0.3092) in Log CFU/g flower tissue between P. protegens 1B1 and P. brassicacearum 93G8 was found 24 hrs after inoculation, however, 72 hrs post treatment application, significantly more Log CFU/g in P. protegens 1B1 were recovered compared to P. brassicacearum 93G8 (p= 0.036) (Figure 3).
Figure 4. Colonization (CFU/g tissue) of ‘Golden Delicious’ apple flowers by Pseudomonas protegens strain 1B1 and P. brassicacearum strain 93G8 immediately after the bacteria were applied to the flowers (>1 hrs) and 24 and 72 hrs post inoculation. Time point >1 hrs represents the initial inoculum load recovered from the flowers. Standard error bars are shown in the graph. Means followed by the same letter at 24 and 72 hrs are not significantly different (p=0.05).
3. Efficacy of Pseudomonas spp. in controlling blossom blight infections
All of the flower clusters treated with Pseudomonas protegens strain 1B1 or P. brassicacearum strain 93G8 were wilted and necrotic, while 80% of the clusters treated with streptomycin were wilted and necrotic (Table 3). All of the clusters that were not treated but challenged with E. amylovora were wilted and necrotic. Amongst the treatments, there was no no significant difference in disease severity with populations exceeding 7.6 Log CFU/g tissue. No disease was observed on the non-treated, non-inoculated control clusters and no E. amylovora was recovered from these clusters.
Table 3. Blossom blight incidence and disease severity following the applications of Pseudomonas protegens strain 1B1, P. brassicacearum strain 93G8 or streptomycin.
| Treatment | No. Clusters Tested | Percent Disease Incidence | Disease Severity (Log CFU/g) |
| Pseudomonas protegens strain 1B1 | 17 | 100 | 8.1 a |
| Pseudomonas protegens strain 93G8 | 17 | 100 | 7.6 a |
| Streptomycin | 15 | 80 | 7.9 a |
| Non-treated, inoculated control | 15 | 100 | 7.9 a |
| Non-treated, non-inoculated control | 6 | 0 | 1 b |
Discussion
Nineteen strains of Pseudomonas spp. tested in this study resulted in inhibitory activity in vitro against four strains of E. amylovora isolated from shoot tissue infected with fire blight. Among these strains, P. protegens strain 1B1 and P. brassicacearum 93G8 were selected to determine their ability to colonize apple blossoms based on their ability to inhibit E. amylovora in vitro (this study) and previously published characteristics of these strains that make them good biocontrol candidates. Pseudomonas protegens strain 1B1 had the highest inhibitory activity against all four E. amylovora strains with an average inhibition zone of 24.2 mm (Figure 2). This strain has also shown moderate inhibition against Rhizoctonia spp. and Pythium spp. (Mavrodi et al. 2012) and high inhibition against Agrobacterium spp. in in vitro studies (Chagas de Freitas, personal communication). Pseudomonas protegens strain 1B1 produces four secondary metabolites known to have bacteriostatic activity – 2,4-diacetylphloroglucinol (DAPG), pyoluteorin, pyrrolnitrin, cyclic lipopeptide, and exoenzymes (Tao et al. 2015; Mavrodi et al. 2012). P. protegens has previously been reported to inhibit the growth of E. amylovora (Yan et al. 2017; Mikicinski et al. 2020). Most recently, Mikicinski et al. (2020) reported that P. protegens strain 59 M (59 M), which originated from soil in Poland, showed zones of inhibition ranging from 0 to 11.5 mm after 48 hr depending on the medium used. In field studies, 59 M reduced blossom infection on cv. Jonagored by 70% compared to the non-treated control. Yan et al. (2017) also confirmed that a strain of P. protegens (pf-5) could inhibit E. amylovora in vitro. They also confirmed that DAPG and pyoluteorin were responsible for the inhibition of E. amylovora.
The second Pseudomonas species selected in this study, P. brassicacearum 93G8, also had inhibitory activity against all four E. amylovora strains with an average inhibition zone of 19.5 mm (Figure 2). Similar to P. protegens strain 1B1, P. brassicacearum 93G8 has shown high inhibition against Agrobacterium spp. in in vitro studies (Chagas de Freitas, personal communication) and produces two of the same bacteriostatic secondary metabolites (DAPG and exoenzymes) (Tao et al. 2015). Unique to P. brassicacearum 93G8 however, is the production of hydrogen cyanide (HCN). Pseudomonas brassicacearum are known biocontrol agents against soilborne pathogens and are responsible for the suppression of take-all disease in monoculture wheat fields (Schlatter et al. 2017) and also can reduce bacterial canker (caused by Clavibacter michiganenesis subsp. michiganenesis; CMM) severity in tomato (Lanteigne et al. 2012; Paulin et al., 2017). Interestingly, Paulin et al. (2017) determined that P. brassicacearum LBUM300 requires both DAPG and HCN production to reduce populations of CMM, but the overexpression of DAPG was primarily responsible for increased colonization of the rhizosphere by LBUM300. DAPG is produced by other species of Pseudomonas, most notably P. fluorescens, and is commonly associated with the inhibition of phytopathogens (Keel et al. 1996). Although the mechanism of inhibition by P. protegens strain 1B1 and P. protegens strain 1B1 were not determined in this study, it is probable that DAPG alone or in combination with pyoluteorin or HCN are the primary mechanisms of inhibition of the Ohio strains of E. amylovora.
One of the hurdles of using bacteria as BCAs is establishing high populations of the bacteria on the target tissue. Ideally, the BCA would be ubiquitous to the target tissue, in this case apple blossoms, and would colonize the tissue and reproduce on the tissue. However, this is not a requirement of an effective BCA. While commercial BCAs are available for managing fire blight, their efficacy in the field is variable and highly dependent on their ability to successfully colonize the blossoms during bloom (Sundin et al. 2009). In this study, when applied at high concentrations to open flowers, 1B1 and 93G8 initially dropped in concentration after 24 hrs but recovered to concentrations equal to the initial inoculum load applied to the blossoms (Log 7.0 and 6.8 respectively). E. amylovora shifts from an epiphytic phase to an infection phase when cell populations reach around 10 000. Consequently, at populations of Log 7.0 and 6.8 cells per gram of tissue, 1B1 and 93G8 are promising BCA candidates. Other antagonistic bacteria, such as Pantoea agglomerans and P. fluorescences strains have also been found to be good colonizers of apple flower stigmas reaching population sizes close to 6 log CFU per flower (Pusey 2002; Wilson and Lindow 1993). Although 1B1 and 93G8 successfully colonized the flowers and inhibited E. amylovora in vitro, they did not prevent E. amylovora infections when tested in planta. In this preliminary study only one application of 1B1 and 93G8 was made. Since E. amylovora can cause infections during the entire bloom period, follow-up studies using multiple applications of 1B1 and 93G8 are warranted.
Educational & Outreach Activities
Participation Summary:
Oral presentation at the 2020 Ohio Produce Network, Ohio Produce Growers & Marketers Association that took place at the Columbus Airport Marriott in Columbus, OH on January 22-23, 2020.
Graduate student exit seminar, The Ohio State University, Dept. Plant Pathology, Wooster, OH April 4, 2021.
Research update at the 2022 Ohio Fruit Growers marketing Association (FGMA) annual winter meeting, February 3, 2022 (Virtual)
Project Outcomes
The results of this study will be used to build on new studies that investigate how best to apply biocontrols in the field and at what frequency they should be applied. In addition, integrating these novel strains of Pseudomonas with other sustainable practices such as host resistance/tolerance, tree spacing and water management will contribute to future sustainability of fire blight management.
As the advisor my knowledge about implementing sustainable practices, specifically biological control in this study, for fire blight management was unchanged. Fire blight is a complex disease caused by the bacterium Erwinia amylovora. Bacterial diseases are notoriously hard to control, especially in perennial crops where they are often systemic. Developing sustainable management strategies that are easily implemented by the grower requires an integrated, systematic approach, which can take years. While we identified strains of Pseudomonas that could inhibit E. amylovora in vitro, they did not inhibit E. amylovora in vitro under the conditions we used in the study. This was not unexpected. Many preliminary studies identify biocontrol organisms that are effective under laboratory settings but not under field settings.