Final report for GNE19-217
Project Information
Alfalfa is the third largest field crop produced in the US, with 2018 hay values calculated over $9.9 billion. However, alfalfa is susceptible to fungal pathogens and drought, which are increasing threats as climate patterns become more extreme and unpredictable. The purpose of this work is to determine if application of the plant growth promoting rhizobacteria (PGPR) Bacillus subtilis UD1022 (‘UD1022’) along with the legume symbiont Sinorhizobium meliloti will enhance alfalfa crop productivity. The PGPR UD1022 has been shown to antagonize common fungal pathogens including Phytotphora capsica, Rhizoctonia solani and Botrytis cinerea and to increase plant biomass in many horticultural plant species. The combination of UD1022 and alfalfa symbiont S. meliloiti could provide protection from pathogens and increase alfalfa yields.
In this project we evaluated the direct effect PGPR UD1022 has on four common fungal pathogens of alfalfa including, Phytophthora medicaginis (root rot), Colletotrichum trifolii (anthranose), Phoma medicaginis (blackstem) and Fusarium oxysporum f. sp. medicaginis (vascular or fusarium wilt). These pathogens have a great economic impact on the production of alfalfa; applications of fungicides represent a cost in time and money in attempting to reduce the effect of the disease and crops impacted suffer loss in yield and nutritional value. To determine if UD1022-produced natural compounds antagonize fungal growth, the bacteria and fungus strains were grown together on petri plates directly or in separate compartments. We found UD1022 to be strongly antagonistic toward three of the four pathogen species tested when grown directly; UD1022 did not influence any of the fungus when grown in divided plates. Initial testing of in planta capability of UD1022 to protect alfalfa from Phytophthora Root Rot were inconclusive.
To evaluate the overall growth promotion effect of inoculating both UD1022 and S. meliloti onto alfalfa, we conducted a pot study using field soil in controlled plant growth chambers. The treatments include no inoculation of bacteria, S. meliloti only, UD1022 only and co-inoculation of both S. meliloti and UD1022. The biomass and nodulation of the plants were compared to observe the response of alfalfa to the application of the PGPR. Based on growth responses, there is encouraging evidence that UD1022 may increase the biomass and number of pink root nodules of alfalfa.
Due to time constraints caused by the suspension of University of Delaware research during Covid-19 laboratory lock-down, drought studies and field studies were discontinued for this project.
- Determine direct fungal antagonism of UD1022 on four fungal pathogens of alfalfa using standard laboratory assays.
1a. Evaluate biofilm and non-ribosomal protein B. subtilis UD1022 mutants for their role in fungal antagonism.
- Test alfalfa (Vernal, susceptible) resistance to Phytophthora medicaginis strain A2A1 oomycete alfalfa pathogen when inoculated with both S. meliloti and UD1022 compared to un-inoculated and S. meliloti alone.
- Quantify alfalfa biomass and number of symbiotic root nodules of plants inoculated with S. meliloti, UD1022, both S. meliloti and UD1022 and no inoculation under growth chamber conditions. Characterize differences in shoot and root biomass and number of nodules between treatments.
The purpose of this project is to enhance alfalfa crop productivity using plant beneficial microbial inoculants. The use of nitrogen (N) fertilizers in agriculture is unsustainable and costly. Leguminous plants, such as soy and alfalfa are known to mitigate these liabilities through their ability to form symbiosis with N-fixing rhizobia bacteria in their roots. Rhizobia symbionts are commonly applied legume crop seed, reducing nitrogen inputs, and enhancing plant growth. However, alfalfa production is susceptible to many fungal pathogens. There is a growing effort in the agro-chemical industry toward the use of natural microbial inoculants to promote the growth of vital food production crops. These ‘bioinoculants’ have been shown to benefit plants through increasing biomass, suppressing bacterial and fungal pathogens, and increasing nutrient uptake. This project will assess the compatibility and capabilities of the University of Delaware patented plant growth promoting rhizobacteria (PGPR) Bacillus subtilis UD1022 (hereafter ‘UD1022’) applied with the alfalfa symbiont Sinorhizobium meliloti strain Rm8530 (hereafter ‘Rm8530’) to enhance alfalfa crop production.
Alfalfa is the third largest crop grown in the United States, with alfalfa hay valued at $9.9 billion annually. In 2017, Pennsylvania maintained 400,000 acres in alfalfa production, rivaling that of New York and nearly 3 times more than 13 other states in the Northeast Region combined. Alfalfa is important as a modifier of nitrogen inputs, both as a cash crop and when used in rotations. Its superior ability to take up and retain nitrogen contribute to its high protein content as fodder and makes it an ideal crop for nitrogen management. UD1022 is documented to increase pea plant biomass and nitrogen content when applied with the pea symbiont. The co-inoculation of UD1022 with S. meliloti could increase alfalfa biomass and its nutritional value.
Fungal pathogens pose a formidable challenge to the productivity of alfalfa production. The PGPR UD1022 has been shown to antagonize common fungal pathogens including Phytotphora capsica, Rhizoctonia solani and Botrytis cinerea, likely through volatile compound production. These and other common fungal pathogens significantly reduce alfalfa crop yields and require intensive use of chemical control. The use of UD1022 in addition to an S. meliloti alfalfa symbiont has the potential to drastically reduce the need for chemical controls and simultaneously provide additional benefits and protections to alfalfa.
Research
1.0 UD1022 Fungal Antagonism Assays
1.1 Direct Antagonism Assay.
Bacillus subtilis UD1022 was used to challenge four common pathogens of alfalfa, obtained from Dr. Deborah Samac (University of Minnesota). The fungal isolates tested were Phytopthora medicaginis (root rot) strain A2A1, Phoma medicaginis (spring black stem rot), strains StC 306-5 and StC 306-10, Fusarium oxysporum f. sp. medicaginis (fusarium wilt) Fom 255 and Fo 3, and Colletotrichum trifolii (anthranose) strains St. Paul AN 1 and SM. Appropriate permitting was obtained through the USDA APHIS program. Fungal isolates were cultured on Potato Dextrose Agar (PDA) and V8 agar (A2A1) and stored on plates. A plug of fungal culture (7 mm) was cored and placed on a petri plate with a 20 microliter drop of UD1022 culture at a distance of 2 cm. Fungal area was measured and compared to controls without UD1022. Three biological replicates were used, and the experiment was repeated three times. Mean fungal areas (cm2) were compared after evaluation of normalcy. Tukey-Kramer test was applied, and significance was accepted at P less than 0.05. This experiment determined UD1022 direct antagonism toward common alfalfa fungal pathogens.
1.2 Indirect Antagonism Assay.
This test was performed similarly to the direct assay, except the petri plate is divided in the middle to prevent direct interaction.
2.0 UD1022 Plant Fungal Suppression Assay.
Alfalfa (Vernal) seed was sterilized and cold stratified overnight at 4 degrees C. Seeds were sown to pots containing a mix of peat:vermiculite:Turface (3:2:1). Plants were maintained in controlled growth chambers with 16-hour photoperiod, 55 percent relative humidity at 22 degrees C. Plants were thinned to 10 per pot and 24 pots are included for each treatment: treatments of no bacteria control, S. meliloti, UD1022 and S. meliloti plus UD1022 were inoculated at time of seed sowing and a ‘booster’ of UD1022 was applied 9 DAP to ‘UD1022’ treatments. 10 mL A2A1 commutated culture was drenched onto plants 10 DAP. One set (12 pots per treatment) was infected, and one set (12 pots) was subjected to ‘mock’ inoculation’ as ‘control’ plants. Results were evaluated 20 days after transplantation (10 days after infection); plants were rated for Phytophthora Root Rot infection using a 5-class scale from 1= healthy to 5= dead. Statistical analysis applied a test for normal distribution and comparison of mean index values using Tukey-Kramer test and significance of P less than 0.05.
3.0 UD1022 – meliloti Alfalfa growth promotion in Greenhouse.
Field soil was collected from an agriculture field cultivated in corn for 18 years on the University of Delaware Research Farm in Newark, DE in October 2019. The top 6 inches of plowed soil was collected, sieved (4 mm), and stored in 4 degrees C cooler. Soil was homogenized 1:1 with vermiculite and used in 3-inch pots for alfalfa growth. Alfalfa (Vernal) seed was sterilized, stored overnight in 4 degrees C, and sown directly in pots, thinned to 1 plant per pot, placed in greenhouse and maintained similar to Objective 2.0. Twenty-one plants were randomly assigned to four treatments: no inoculation, S. meliloti only, UD1022 only, and S. meliloti with UD1022; bacteria were inoculated same day as sowing. At 4 weeks after planting, 10 plants were randomly selected from each treatment, and UD1022 was re-inoculated creating a total of 8 treatments (4-original treatments and 4-original treatments + “UD1022 Booster”. Plants were harvested 7 weeks after sowing, nodules collected and preserved, roots and shoots separated, and tissues dried at 70 degrees C for 72 hours. Experiment was repeated three times. Shoot material will be sent for nitrate analysis; total nitrogen and total carbon will be evaluated depending on quantity of material available. Data were analyzed by mean comparisons and t-Tests applied with significant differences determined at P less than 0.05.
1.0 UD1022 Fungal Antagonism Assays
1.1 Direct Antagonism Assay.
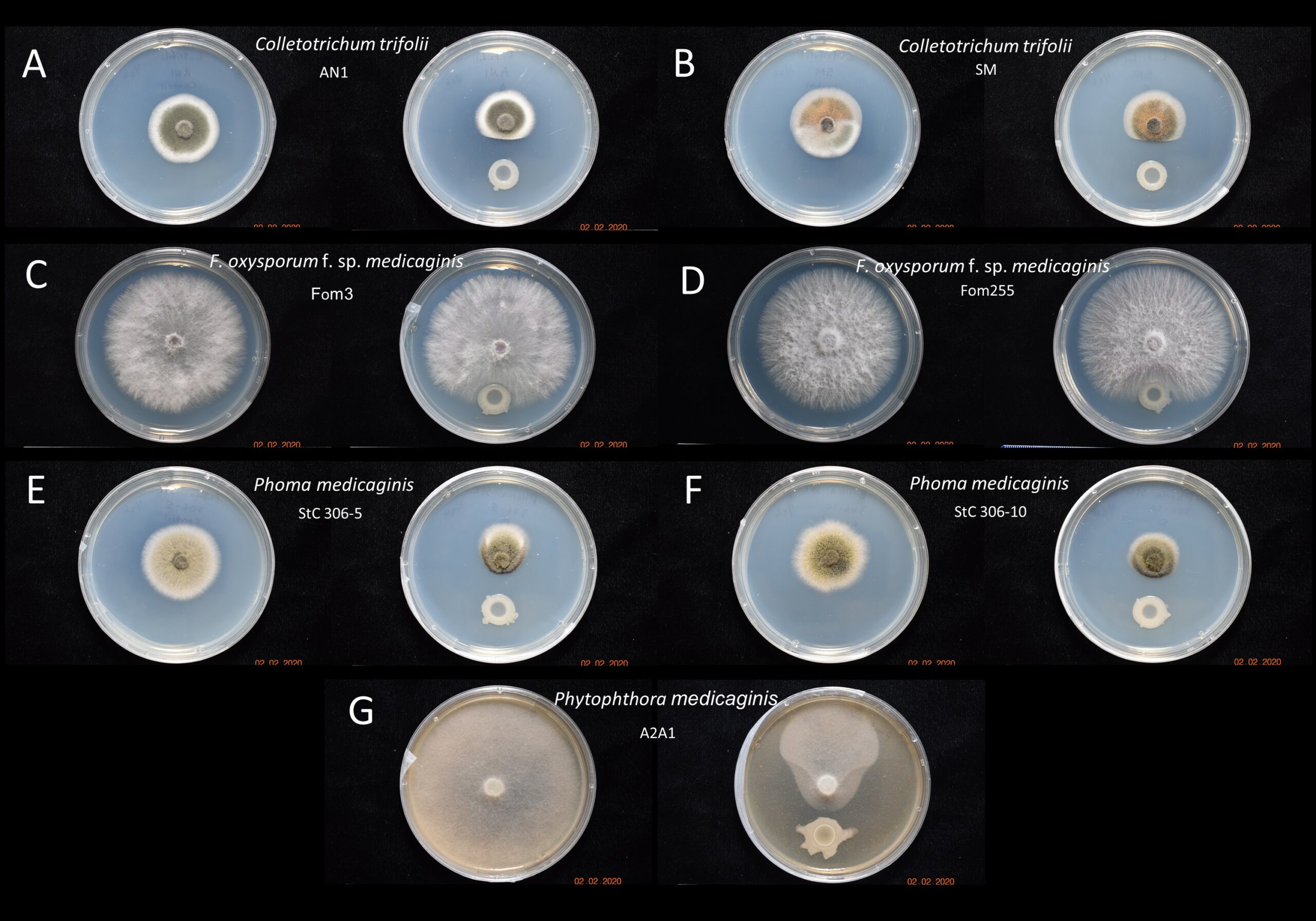
Results suggest that PGPR B. subtilis UD1022 maintained moderate antagonism toward both strains of Colletotrichum trifolii, no antagonism activity against either strain of Fusarium oxysporum f. sp. medicaginis and significant antagonism activity against both strains of Phoma medicaninis and Phytophthora medicaginis. These results indicate that UD1022 could be useful as an alfalfa crop protectant against fungal pathogens such as anthracnose, spring black stem rot and root rot (Figure 1).
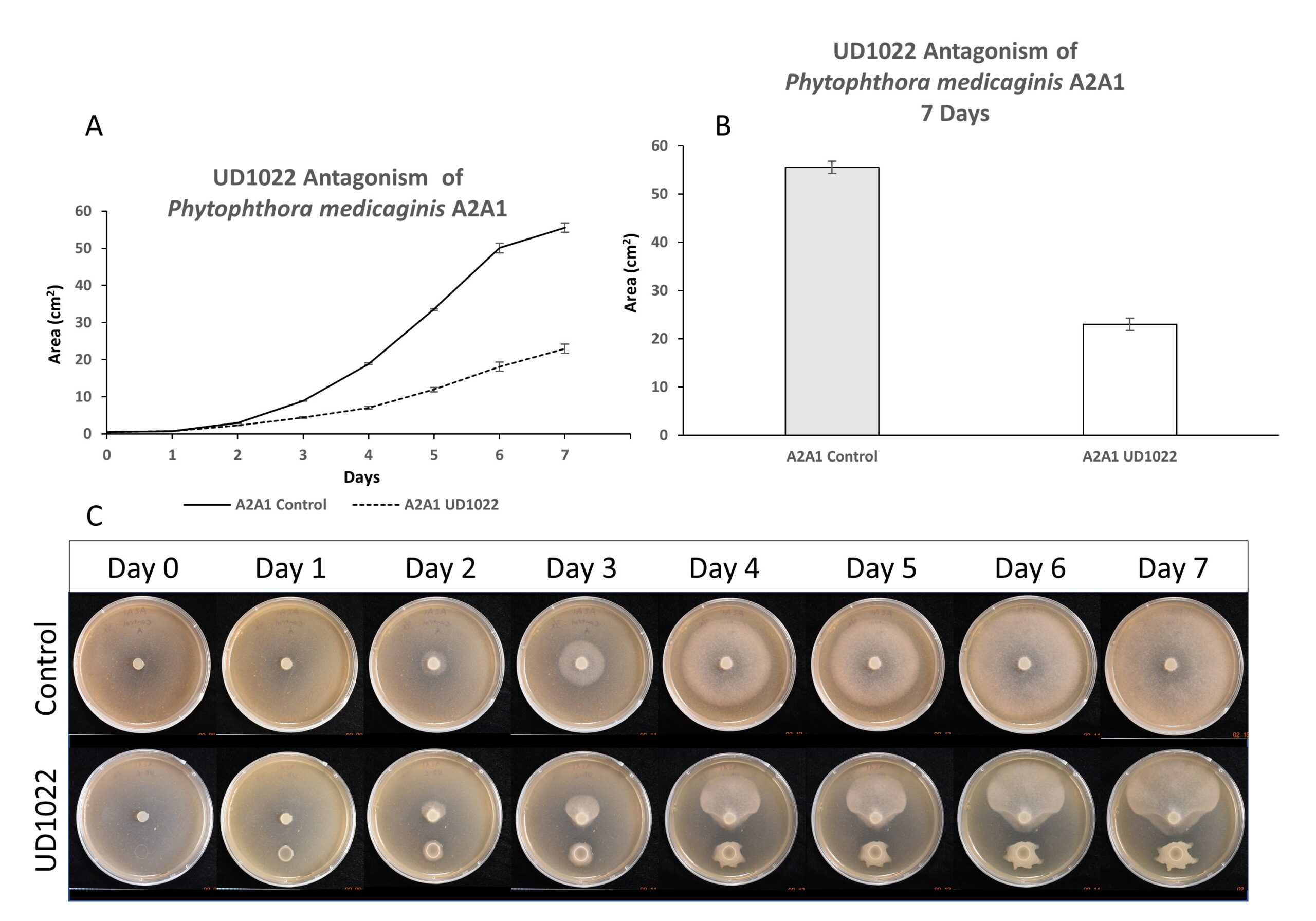
Antagonism of UD1022 toward P. medicaginis strain A2A1 was evaluated over a time course of 7 days; robust antagonism effects were observed after 3-4 days of growth and increased throughout (Figure 2). At 7 days, A2A1 growth was antagonized by 64%.
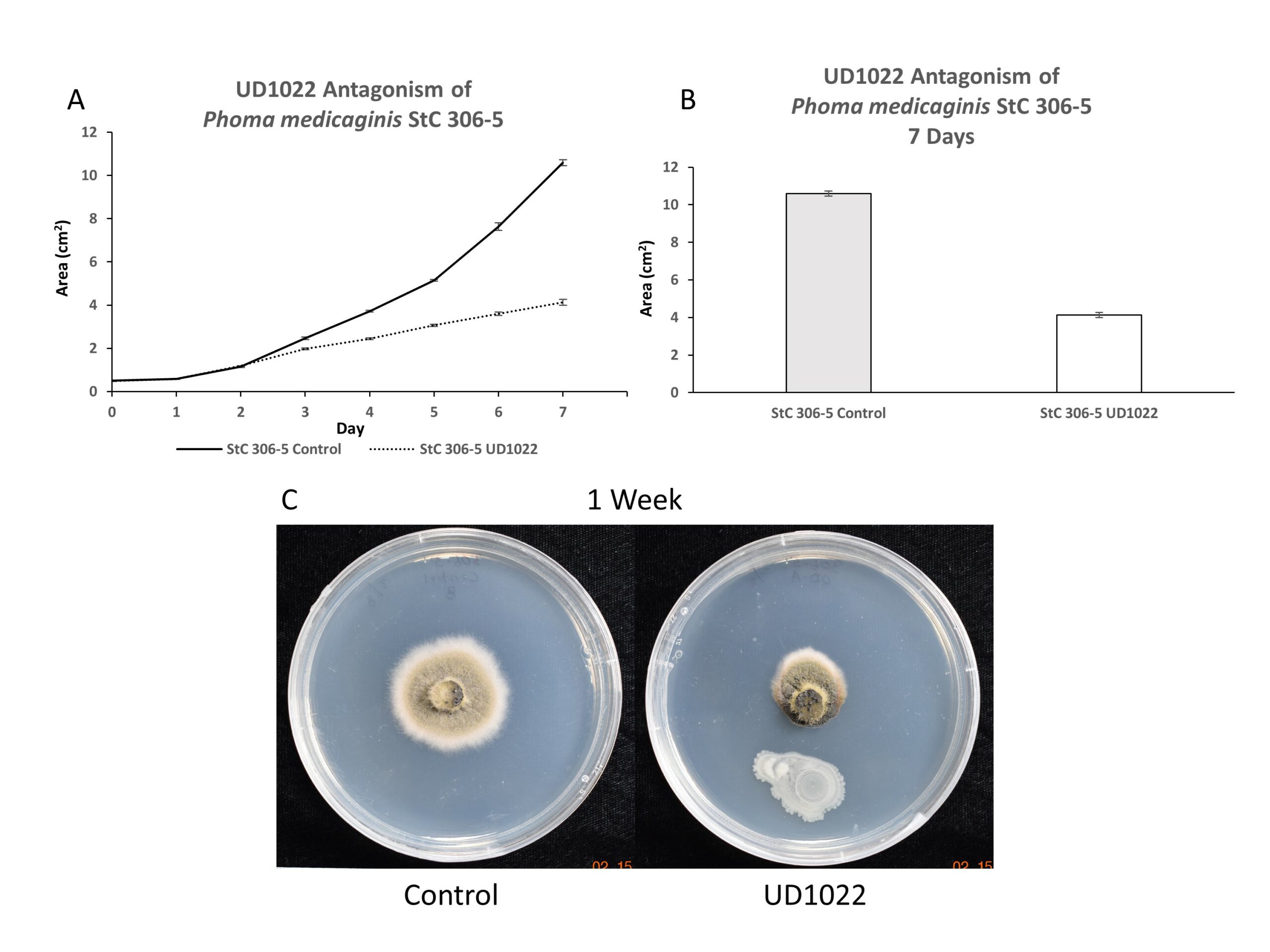
Antagonism of P. medicaginis strain StC 306-5 by UD1022 was evaluated over 7-day time course; antagonism was observed after 4-5 days growth and increased though 7-days. At 7 days, StC 306-5 growth was antagonized by 61%. Appearance of the visible mycelium is darkened and highly impacted compared to control (Figure 3).
1.1a Addendum/Expansion of Direct Fungal Antagonism Analysis.
Several UD1022 mutant strains ineffective in selected non-ribosomal peptides and biofilm genes were used in direct fungal challenge assays against P. medicaginis A2A1 and P. medicaginis StC306-5 to investigate potential mechanisms of UD1022 antagonism.
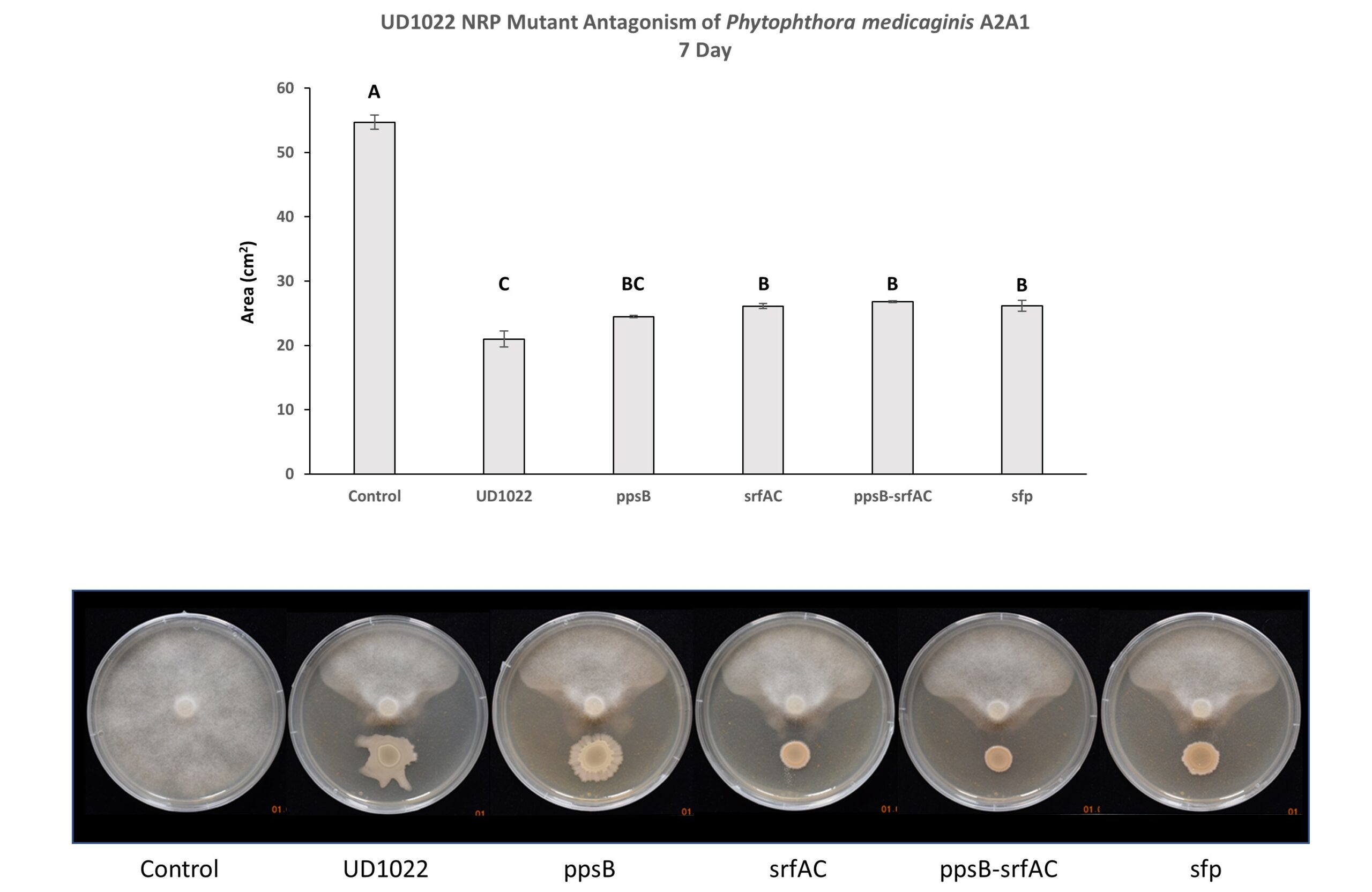
Non-ribosomal peptides (NRP) including surfactin and plipastatin have been implicated in the antagonism of common plant fungal pathogens. UD1022 plipastatin (ppsB), surfactin (srfAC), surfactin critical gene sfp potentially do not contribute to antagonism of A2A1 (Figure 4). Though A2A1 grown with srfAC, sfp, and double mutant ppsB-srfAC were statistically less affected by UD1022 however, they were clearly antagonized by the presence of the mutants. Three separate experiments with 3-technical replicates per treatment within each experiment were performed.
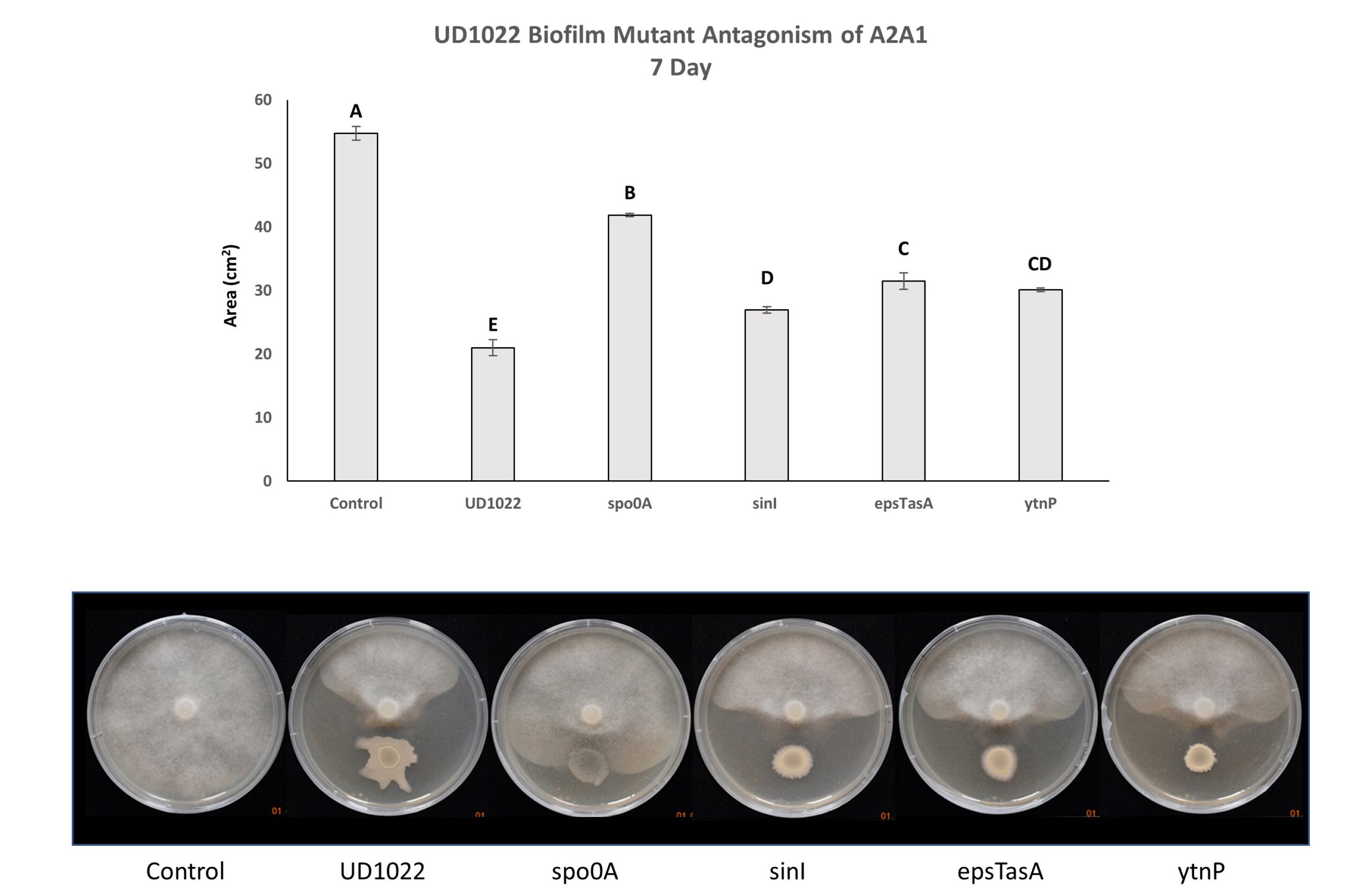
Figure 5. UD1022 Biofilm Mutant Antagonism of P. medicaginis A2A1. The master regulator gene spo0A is potentially required for UD1022 antagonism. B. subtilis biofilm genes sinI and epsTasA may be partially engaged in UD1022 antagonistic activity. The gene ytnP encodes a metalo-lactamase implicated in quorum quenching of other soil bacterial species; it is intriguing that this mutant shows significantly less antagonism.
UD1022 genes responsible for biofilm and spore formation (epsTasA, spo0A and sinI) are potentially necessary to variable degrees for fungal antagonism. Spo0A, a master regulator controlling both biofilm and sporulation pathways in B. subtilis, is significantly different from control and is likely required for antagonism of A2A1. Interestingly, the gene encoding the metalo-lactamase protein YtnP (which cleaves N-acyl homoserine lactones [AHLs]) may have a role in antagonism of P. medicaginis A2A1 (Figure 5). To the authors knowledge Phytophthora spp. have not been reported to produce AHLs; our lab found no detectable AHLs produced by A2A1 using the Agrobacterium tumefaciens reporter strain KYC55/pJZ410/pJZ384/pJZ372 (data not shown). Three separate experiments with 3-technical replicates per treatment within each experiment were performed.
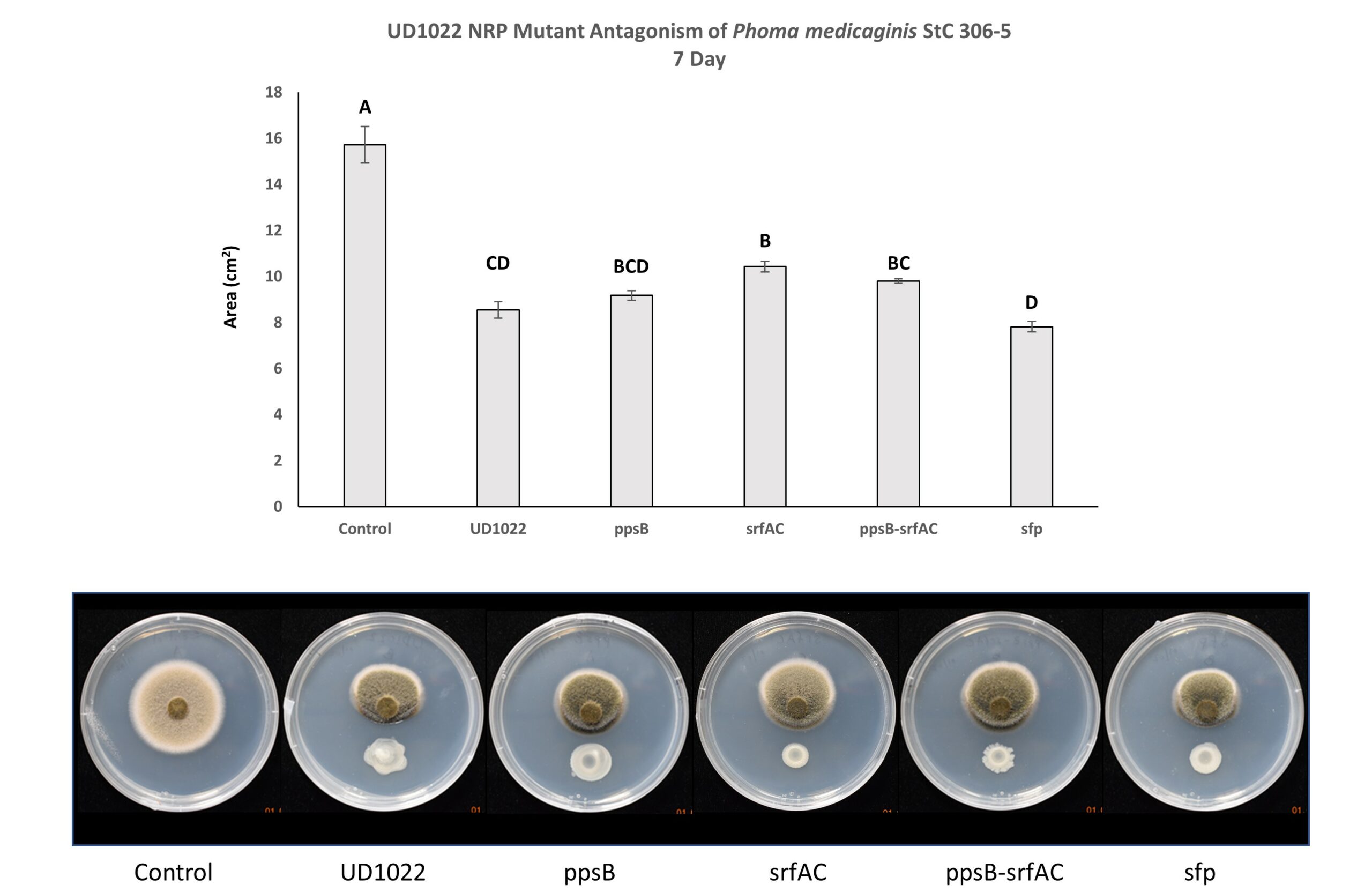
Non-ribosomal peptides (NRP) including surfactin and plipastatin have been implicated in the antagonism of common plant fungal pathogens. UD1022 plipastatin (ppsB), surfactin (srfAC), surfactin critical gene sfp possibly do not contribute significantly to antagonism of P. medicaginis StC 306-5. The lack of the gene srfAC, responsible for producing surfactin, resulted in significantly less antagonism than UD1022; however, this does not alleviate antagonism to the level of control. Interestingly, culturing with the srf mutant consistently resulted in visually greater antagonism than UD1022 WT yet fungal area was not significantly different (Figure 6). Three separate experiments with 3-technical replicates per treatment within each experiment were performed.
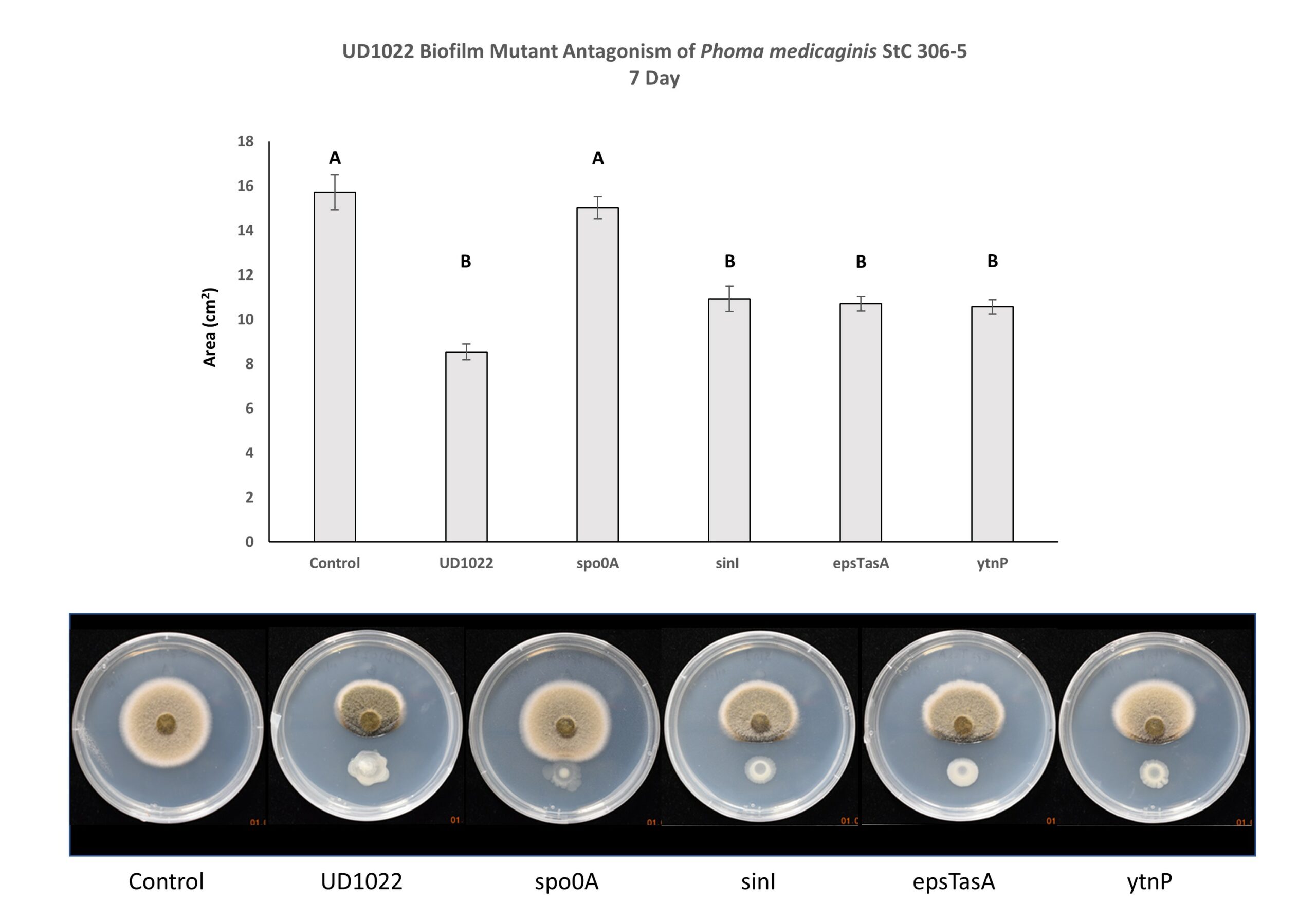
UD1022 genes responsible for biofilm formation (epsTasA and sinI) and lactonase gene ytnP do not appear necessary for fungal antagonism. The fungal area of the Spo0A treatment is significantly different from UD1022 WT, and statistically similar to unchallenged control. Spo0A is strongly implicated in the antagonism of P. medicaginis StC 306-5. (Figure 7).
1.2 Indirect Antagonism Assay
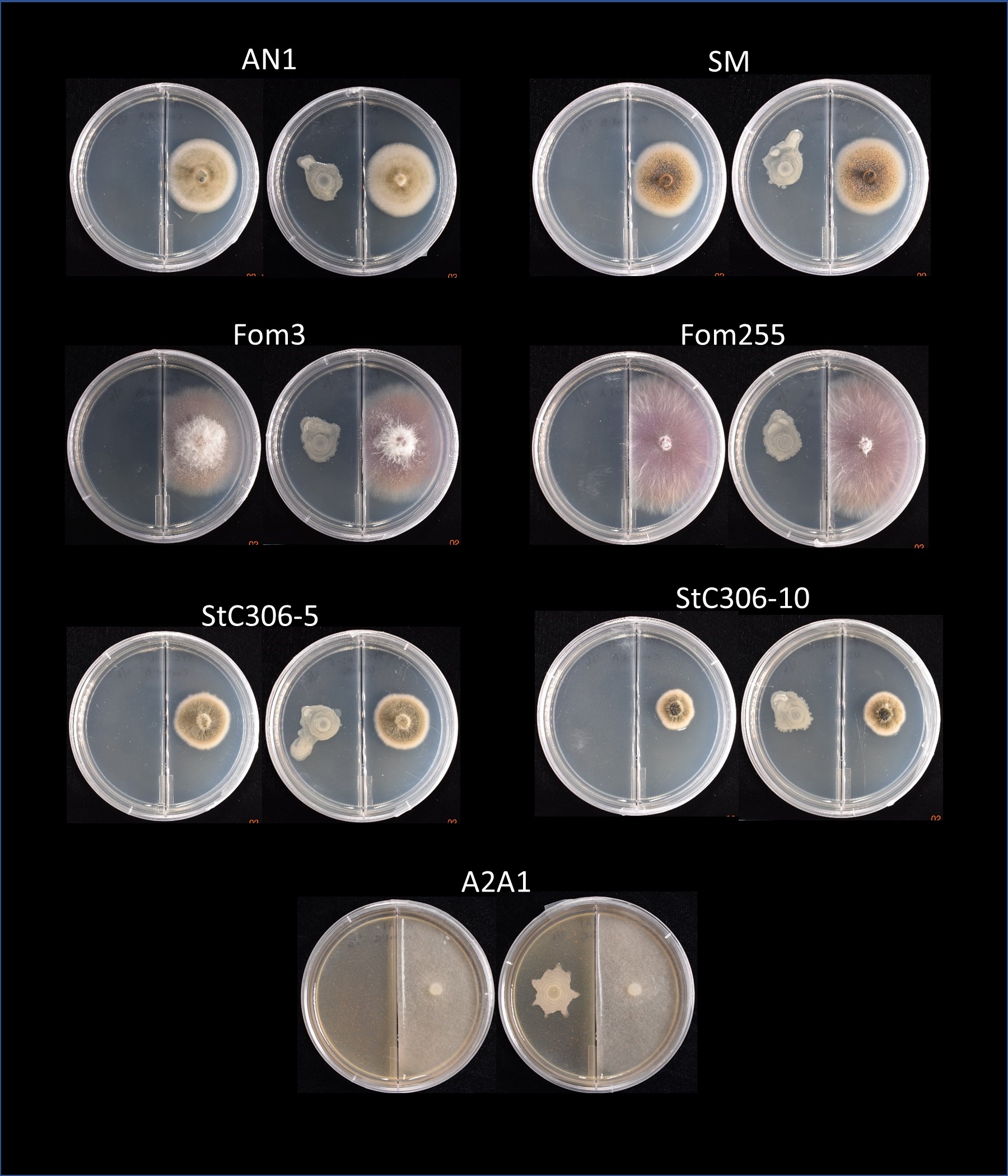
UD1022 produces volatile compounds which may inhibit or alter the growth of other organisms. However, we found no antagonism or inhibition of growth of any of the fungal strains tested when UD1022 was not in direct contact with the pathogen (Figure 8).
2.0 UD1022 In Vivo Fungal Suppression Assay.
In vivo tests of UD1022 suppression of A2A1 on alfalfa (Vernal) in plant under growth chamber conditions were inconclusive potentially due to a variety of factors. Three separate replicates were performed. Disease severity was mild and not visually different from positive and negative controls or treatments for all replicates. Plants inoculated with S. meliloti Rm8530 were unevenly colonized, and plants were not supplemented with nitrogen. Plants overall appeared slightly chlorotic regardless of infection treatment or control.
3.0 UD1022 – S. meliloti Alfalfa Growth Promotion in Greenhouse.
Three replicates of this experiment were completed; shoot and root dry weights and nodule numbers were recorded. Experiment replicate number 1 was dissimilar from 2 and 3 and results were inconclusive. Results from the 2nd replicate test are presented as they corroborate 3rd replicate test results. Statistical analysis suggests that all data fit criteria for normalcy and homogeneity.

Alfalfa plants treated with Rm8530 only, in general, appeared moderately less robust than any of the co-inoculated treatments. However, height and root density were similar to plants in the Rm8530-UD1022 co-inoculation treatment. Individual plants within treatments were not uniform in height; at time of harvest, plants were visually sorted from shortest to tallest, and the middle individual was selected for photographic documentation (Figure 9).
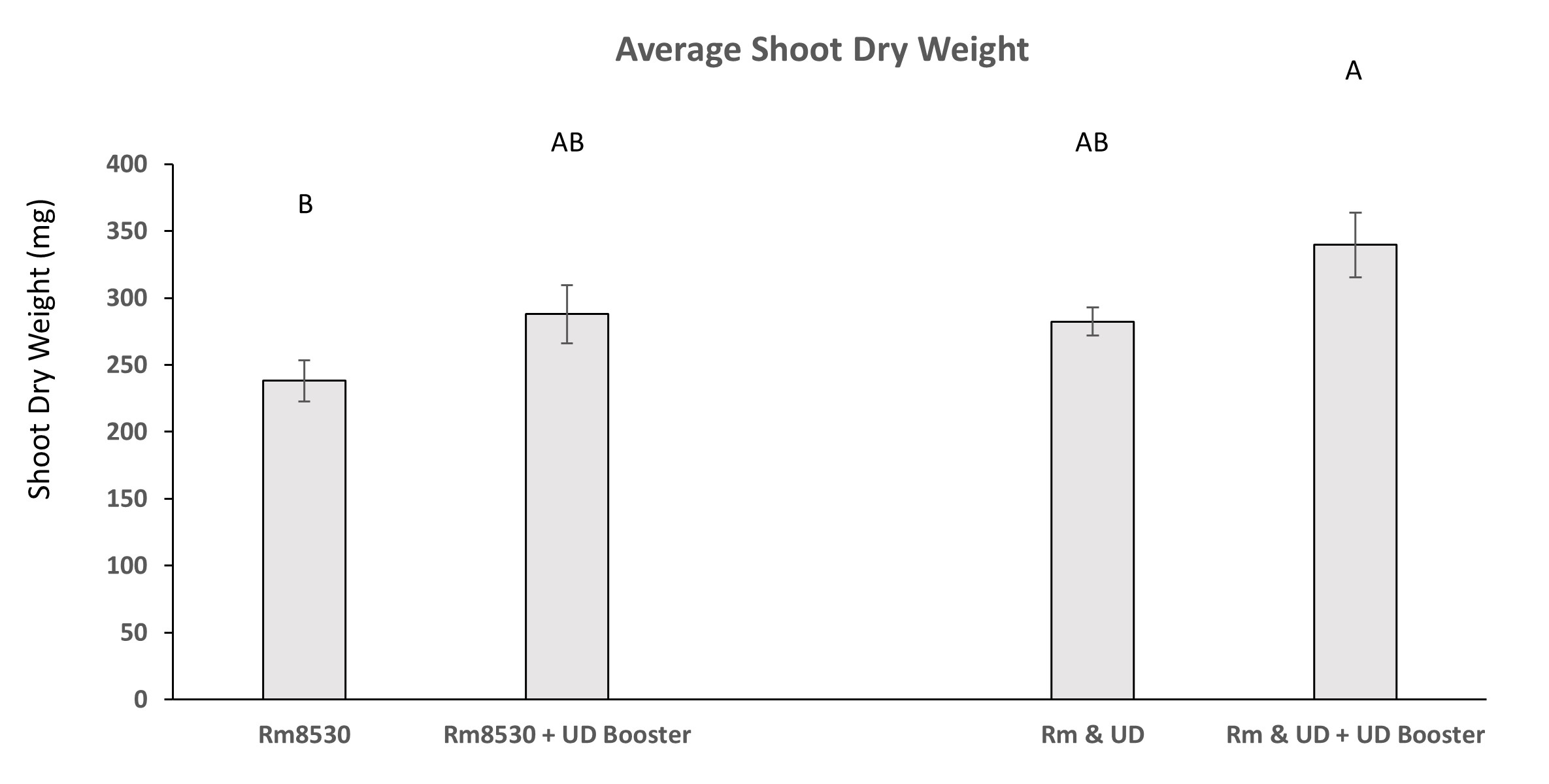
Treatments of alfalfa with the PGPR UD1022 slightly increased shoot biomass when applied at the same time as the rhizobia symbiont Rm8530 and when applied 4-weeks after Rm8530 as compared to treatment with Rm8530 alone. Average plant biomass of alfalfa initially co-inoculated with Rm8530 and UD1022 and re-inoculated with UD1022 after 4-weeks is significantly greater than plants inoculated with Rm8530 alone (Figure 10). Time constraints prevented the analysis of nitrate content in shoots and biomass quantity restraints negated the analysis of total nitrogen and carbon.
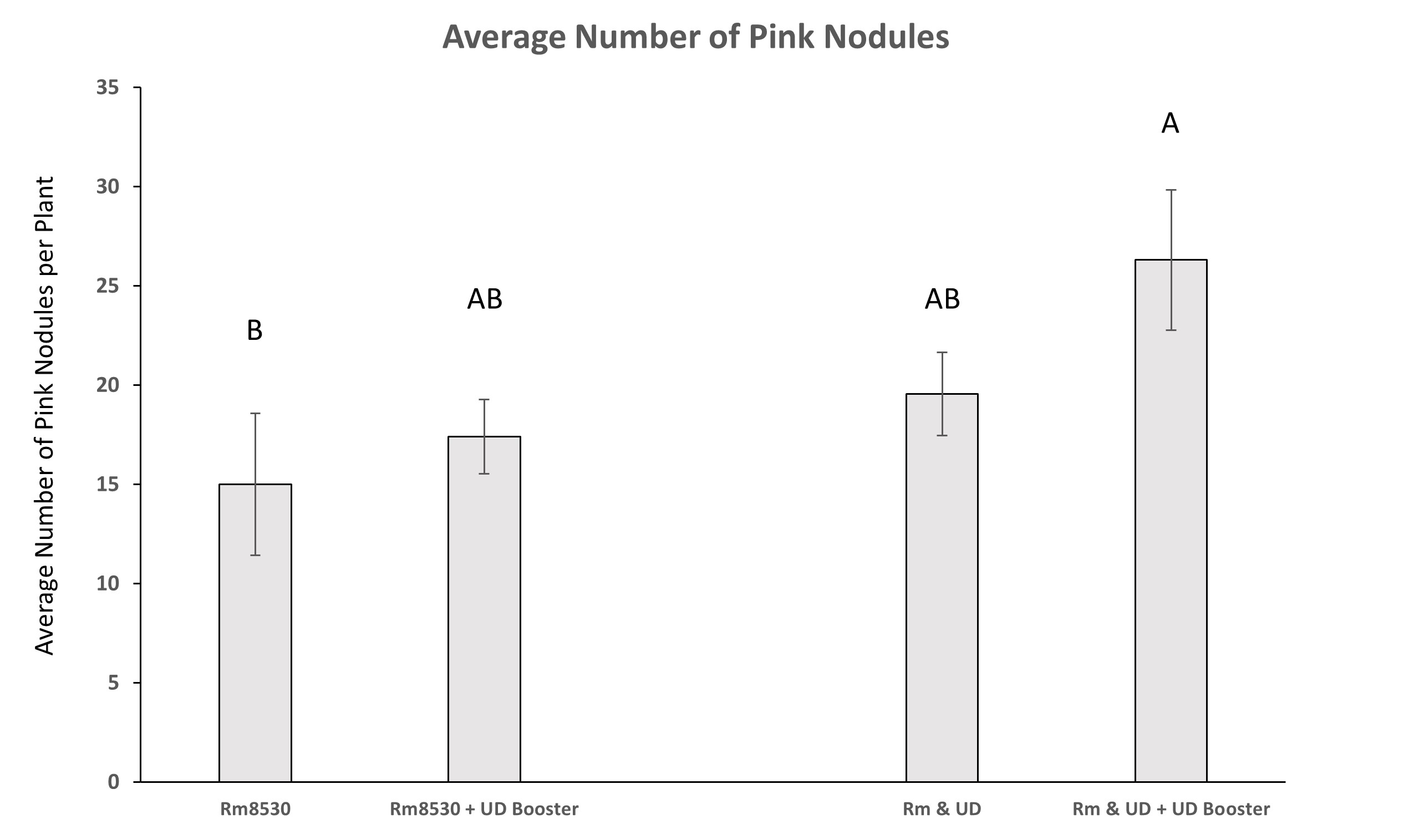
As observed in the shoot biomass results, co-inoculation treatments of UD1022, both at time of planting and after 4-weeks, did not result in significantly greater number of pink nodules over that of Rm8530 alone. Only the treatment of UD1022 ‘booster’ application in addition to the initial co-inoculation of bacteria had significantly greater numbers of pink nodules compared to Rm8530 treatment (Figure 11).
Root biomass as characterized by dry weight were extremely variable between experimental treatments and the data did not fit parameters for normalcy or homogeneity. Growth of alfalfa in field soil presented some challenges, possibly due to the endemic bacterial and fungal populations. This may explain the variability of results between experimental replicates.
The results of the in vitro fungal antagonism assays provide interesting directions for researching mechanisms of UD1022 antagonism toward these two alfalfa pathogens. Phytophthora Root Rot in alfalfa dramatically reduces productivity. Given that the causal organism is an oomycete, control can be challenging in wet conditions. Resistant cultivars of alfalfa have been developed; however, resistance can be genetically variable across stands. Utilization of a biological control approach would be beneficial as an additional tool to manage potential pathogen infection. PGPR UD1022 has shown strong antagonistic effects on P. medicaginis strain A2A1. Though the non-ribosomal proteins surfactin and plipastatin production mutants tested showed no role in this antagonism, additional Bacillus NRPs could be involved. Additionally, we did not test direct application of surfactin or plipastatin. Interestingly, a mutant in the putative quorum quenching (QQ) gene ytnP was observed to be less antagonistic than UD1022 WT. Other work by this lab is investigating the possible mechanisms and effects of UD1022 QQ on other beneficial bacteria in legume rhizospheres. Though we did not detect any quorum sensing molecules (substrate of the YtnP protein) produced by A2A1, the action of the lactonase protein expressed may be required for a yet unknown pathway/product contributing to the antagonism of this oomycete. Our study demonstrates the potential role of biofilm genes, especially spo0A, in UD1022 antagonism toward A2A1. Spo0A is known to be a master regulator switch between biofilm and sporulation pathways in B. subtilis; this ‘all encompassing’ activity may pose a challenge in isolating which functions of the defined pathways are responsible for the antagonism observed.
Spring Black Stem, caused by Phoma medicaginis, is known as one of the most destructive alfalfa diseases, causing both yield loss and reduction in forage quality. There are currently few ideal management options as there are no highly resistant cultivars. UD1022 strongly antagonizes P. medicaginis strain StC 306-5. Though NRP pathway mutant strains of UD1022 were generally still antagonistic toward StC 306-5, the srfAC mutant was visibly and statistically less antagonistic. Follow up experiments challenging StC 306-5 directly with surfactin would be of significant interest. As observed in the Phytophthora assays, the spo0A mutant is completely lacking in antagonism toward this fungus. Understanding the genes and mechanisms of UD1022 antagonism of these important alfalfa pathogens would contribute greatly to developing methods to mitigate these economically devastating plant disease agents.
The in vitro assays of UD1022 treated alfalfa challenged with P. medicaginis A2A2 were inconclusive. This data would have been informative for the real-world potential of UD1022 to be applied for the purpose of protection from Phytophthora Root Rot. It is possible that continuous culturing of the P. medicaginis A2A1 strain on V8 agar altered or reduced its pathogenicity/infectivity. Further work evaluating cytological infection dynamics of A2A1 zoospores on M. sativa Vernal (susceptible) roots colonized by UD1022 may elucidate the cause of our ambiguous results.
UD1022 does show modest potential for plant growth promotion in alfalfa. Alfalfa cultivation confronts multiple challenges related to both biological and climate related threats. The PGPR UD1022 conferred greatest benefits when applied twice; initially at time of sowing and inoculation with the rhizobia symbiont, and after 4-weeks of growth. However, this type of application approach may be an impediment to farmer adoption due to the inconvenience and potential costs associated with multiple passes across a field. Though not measured, it was observed that using field soil resulted in heavy damping off pressure on seedlings treated with neither Rm8530 nor UD1022. Presence of plant beneficial bacteria may play a role directly or indirectly in alfalfa seedling resistance to soil borne agents of damping off syndrome. The second application of UD1022 may have served the plant through one or several other proposed modes of plant growth promotion, including increasing access to nutrients or stimulating plant hormone production or systemic resistance.
Given our current findings, there is potential for UD1022 as a bioinoculant for alfalfa crops. UD1022 shows robust antagonism toward multiple different economically devastating alfalfa pathogens. Application to a notably susceptible cultivar of alfalfa showed positive and generally significant improvements to overall shoot biomass and nodule number, as well as potentially protecting vulnerable seedlings. These results and observations provide strong evidence for further investigations of the use of UD1022 in either larger greenhouse studies or possibly a multi-year field study to evaluate plant growth promotion. Specific fungal antagonistic or suppressive activity should also be evaluated using greater quantifiable approaches rather than qualitative disease indexing. Cytological and microscopic approaches coupled with further investigations of the molecular genetic mechanisms of UD1022 fungal interactions could pinpoint specific targets which could be utilized in microbial inoculants for alfalfa crops.
Education & Outreach Activities and Participation Summary
Participation Summary:
Conferences and opportunities to share results of research have been severely impacted by the Covid-19 pandemic. Researchers will be actively looking for forums where the findings of this work may be communicated to the broader community.
Additional outreach will include presenting results from in vitro and in vivo fungal inhibition assays and growth promotion data may be presented in poster/short talk form at the 2022 North American Alfalfa Improvement Conference (NAAIC) held in Lansing, MI.
Project Outcomes
The results of this project are a valuable proof-of-concept strongly supporting research scaling up the application of UD1022 to multi-year field trials of alfalfa.
Given our current findings, there is potential for UD1022 as a bio-inoculant for alfalfa crops. UD1022 shows robust antagonism toward multiple different economically devastating alfalfa pathogens. Application of UD1022 to a notably susceptible cultivar of alfalfa showed positive and generally significant improvements to overall shoot biomass and nodule number, as well as potentially protecting vulnerable seedlings.
These results and observations provide strong evidence for further investigations of the use of UD1022 in either larger greenhouse studies or possibly a multi-year field study to evaluate plant growth promotion. Specific fungal antagonistic or suppressive activity should also be evaluated using greater quantifiable approaches rather than qualitative disease indexing. Cytological and microscopic approaches coupled with further investigations of the molecular genetic mechanisms of UD1022 fungal interactions could pinpoint specific targets which could be utilized in microbial inoculants for alfalfa crops.