Final report for GNE21-261
Project Information
Salmonella enterica serovar Dublin (S. Dublin) is a bovine-adapted emerging pathogen in the northeastern United States. The disease presentation is nonclassical in cattle, particularly in calves, and complicates diagnosis. S. Dublin detection from environmental samples is challenging because the pathogen is often outcompeted by the background flora. Due to persistent host colonization, unclassical presentation, detection difficulty, and treatment limitations arising from antimicrobial resistance, maintaining health of dairy herds is largely dependent on preventative risk management. The epidemiology and effects of S. Dublin on dairy production in the eastern United States are under-researched and, thus, poorly understood. Because Pennsylvania is ranked 7th nationally for dairy production, and the cattle trade spans many northeastern states there is an urgent need to address these knowledge gaps surrounding prevalence and provide data-driven directions for S. Dublin mitigation.
To this end, we collaborated with partners in Penn State Extension and with the Lancaster Dairy Herd Improvement Association to conduct bulk tank milk testing for antibody against S. Dublin, an indicator of infection. Producers with positive samples were given the option for follow-up environmental sampling to isolate S. Dublin from their farms. Additionally, as a possible alternative to antimicrobial treatment, we collected wastewater and manure lagoon samples from which we aimed to isolate bacteriophages against S. Dublin. Beyond research activities, we sought to raise awareness of this issue among farmers in Pennsylvania and accomplished this via informational pamphlets distributed at Penn State’s Ag Progress Days, an article summarizing the topic published through Penn State Extension, and a presentation given to clinicians at the Mid-Atlantic States Conference for Bovine Practitioners.
Through milk testing of 72 farms, we were able to identify four with positive samples. Of these, two elected follow-up environmental sampling. From these samples, we isolated 23 Salmonella sp. strains; however, none were serotyped as S. Dublin. Additionally, no bacteriophages against S. Dublin were present in the environmental samples that were screened. These results do not preclude the presence of S. Dublin in the environment on these farms, but rather point to some of the known challenges and potential biases of culture-based detection of different Salmonella serovars.
Collectively, this project was an important first step toward understanding the prevalence of S. Dublin in Pennsylvania. We confirmed that farms are in fact being impacted by S. Dublin which will inform further research. Moreover, this lays the groundwork for needed awareness among farms and clinicians in the state. The networks established throughout this project will facilitate future education and outreach, thereby better equipping all stakeholders with the knowledge needed for maintain the health and production performance of dairy cattle in Pennsylvania.
Objective 1: Determine prevalence of Salmonella Dublin on dairy farms in Pennsylvania
While the ideal scenario would be to repeatedly sample all PA dairy farms on three separate occasions for accurate determination of the prevalence of S. Dublin, such experimental design is beyond this project’s scope. Thus, we will perform ELISA antibody tests at 900 randomly selected PA dairy farms to estimate the seroprevalence of Salmonella Dublin in the state. This initial screen will yield multiple outcomes. First, it will confirm the presence of Salmonella Dublin in PA dairy operations. The number of positive farms will provide an estimate of prevalence in the state. This knowledge will inform dairy producers of the need to implement risk management intervention strategies as applicable. Finally, fulfilling this objective will allow us to streamline downstream environmental sampling to only those farms seropositive for Salmonella Dublin.
Objective 2: Confirm active presence of Salmonella Dublin on farms with antibody-positive milk
Given the challenging nature of culturing Salmonella Dublin from environmental samples, selective enrichment and PCR-based testing will be used to determine its prevalence on 10 seropositive dairy farms. Based on the work of Cummings et al. 20184 in New York, we expect 0.9% of farms to be seropositive; however, we are in communication with PSU extension team and Penn State Diagnostic laboratory concerning Salmonella Dublin and will target farms that previously tested positive for this pathogen for environmental sampling of manure and calving area. Fulfillment of this objective will confirm active presence of Salmonella Dublin on dairy farms in PA.
Objective 3: Isolate and screen bacteriophages for potential therapeutic intervention
Salmonella Dublin is bovine-adapted and largely, multidrug resistant to antibiotics. In fact, most S. Dublin isolates are resistant to all antimicrobials labeled for treatment of respiratory infections in bovines in the US.2 These characteristics, in combination with general trends toward antimicrobial resistance, justify the need for alternative therapeutic interventions. Phage therapy is increasingly appreciated for its efficacy against multidrug resistant infections in human and animal hosts. Given the resistance of Salmonella Dublin to traditional intervention and the limitations of current preventative measures, phage therapy has the potential to complement existing interventions and improve animal health and production outcomes. We will isolate phages from environmental reservoirs and perform screening for possible candidate phages. This screen will identify phages targeting Salmonella Dublin, providing a phage population that can be further narrowed based on established phage therapy criteria.
The purpose of this project was to estimate the prevalence of Salmonella Dublin in Pennsylvania dairy herds and provide intervention directions to dairy producers facing challenges with this pathogen. Salmonella enterica serovar Dublin (S. Dublin) is a bovine-adapted emerging pathogen in the northeastern United States. The disease presentation is nonclassical in cattle, particularly in calves, and complicates diagnosis.1,2 S. Dublin detection from environmental samples is challenging because the pathogen is often outcompeted by the background flora.3 Due to persistent host colonization, unclassical presentation, detection difficulty, and treatment limitations, maintaining health of dairy herds is largely dependent on preventative risk management.
The epidemiology and effects of S. Dublin on dairy production in the eastern United States are under-researched and, thus, poorly understood. To date, only one study of S. Dublin prevalence among dairy herds in New York has been published.4 Because Pennsylvania is ranked 7th nationally for dairy production5, and the cattle trade spans many northeastern states there is an urgent need to address these knowledge gaps surrounding prevalence and provide data-driven directions for S. Dublin mitigation.
Antimicrobial resistance (AMR) of S. Dublin is also a significant concern, and most farm isolates are multidrug-resistant (MDR),1,6,7,8 creating demand for strategies that prevent infection as well as more sustainable alternative therapeutics.9,10,11 This project will gather preliminary knowledge for the potential development of an alternative to antimicrobials, phage therapy, which is increasingly studied and recognized for its efficacy in treating MDR infections in humans and animals.11,12,13 Because bacteriophages have high host specificity, phage therapy does not apply the broad spectrum selective pressure for AMR of traditional antibiotics. While resistance development is possible, this can be circumvented by applying a combination of phages, known as a phage cocktail, to mitigate resistance development. However, when bacteria develop resistance to phages, there is an evolutionary cost associated with it. For example, in a hypothetical scenario where phage therapy is applied to treat MDR bacteria and phage resistance arises, pathogens usually become susceptible to antimicrobials that they were previously resistant to or become more easily killed by an animal’s immune system.12 Thus, either as a cocktail or in combination with a weaker antibiotic, phage therapy could be effective for treating MDR S. Dublin infections.
This project addresses the emergence of S. Dublin in the northeastern US and investigates potential intervention strategies. The results gathered here will increase awareness among PA farmers concerning the presence of Dublin within their operations and provide preliminary direction for an alternative to antimicrobials. Our results will contribute to reducing animal loss and the need for antimicrobial intervention, leading to a more sustainable dairy production approach. This initial step toward phage therapy is significant because it has the potential to work in tandem with lower dose antibiotics, decreasing their demand and thus relieving some selective pressure toward AMR. The immediate implications of this project are early risk assessment and management, while longer-term implications include potentially reducing animal loss and broad-spectrum antibiotic use in PA dairy production.
Cooperators
- (Researcher)
Research
Objective 1: Determine prevalence of Salmonella Dublin on dairy farms in Pennsylvania
1.1 Participant Recruitment and Sampling
Via collaboration with university and extension partners, we identified Pennsylvania dairy farms willing to participate. Following communication with the Lancaster Dairy Herd Improvement Association (DHIA) testing labs and milk co-op consent, bulk tank samples collected through the DHIA's routine testing were obtained. Additionally, samples were collected from individual farms that enrolled following communication with Penn State extension partners. Without coordination with DHIA and extension, farms were generally unresponsive to recruitment efforts. As a result, study participation relied on a convenience sample of any farm willing to participate following communication with extension veterinarians, extension partners, or DHIA.
1.2 Seroprevalence Screen
In accordance with PrioCHECK Salmonella Ab bovine Dublin protocol, 100 mL of each raw milk sample were used in each of two technical replicates in an indirect enzyme-linked immunosorbent assay (ELISA) against the O antigen of S. Dublin. Results of these colorimetric reactions will be read at 450nm using spectrophotometry. Samples containing antibody against S. Dublin will react in the provided kit reagents, resulting in a color change reflected by OD450 values. These results will be compared to those of provided positive controls and will be corrected using the provided negatived control. Percent positivity will be calculated, with > 35% as the positivity threshold.
1.3 Reporting/Follow-Up
All farms were notified of their test result either directly or via the Lancaster DHIA if the sample was obtained there. The farms with seropositive samples were notified of the option for additional environmental sampling for objectives 2 and 3.
Anticipated Objective Outcomes:
Determining estimated S. Dublin seroprevalence among dairy herds in Pennsylvania
Objective 2: Confirm active presence of Salmonella Dublin on farms with antibody-positive milk
2.1 Environmental Sampling
For farms with seropositive samples that opted for follow-up environmental sample, the following procedure was followed.
Environmental samples were aseptically collected as gauze swabs of different surfaces around the maternity pen, group calf housing, and milkhouse. According to the protocol in Goodman et al.,3 these swabs were placed in a sterile Whirl-pak bag with ~5 mL of sterile evaporated skim milk and stored on ice or cold packs for transport to maintain bacteria viability.
2.2 Confirmatory Subculturing and Characterization
Using the improved protocol for Salmonella spp. detection by Goodman et al.,3 45 mL of RVS broth was added to the Whirl-pak bags (2.1) and incubated for 20-24 hours at 40-44°C. As outlined, subculturing was performed on all samples by plating onto xylose-lysine-deoxycholate (XLD) agar plates and incubated overnight at 33-37°C. Uniform black colonies were then selected for confirmatory molecular identification.
2.3 Molecular Diagnostics
To confirm isolates as Salmonella prior to subtyping, invA PCR is used to confirm a putative bacterial isolate as Salmonella. Using the S. Dublin specific PCR primers developed by Afroj et al (2017), isolates were typed as S. Dublin or not.
Anticipated Objective Outcomes:
Confirmation of estimated S. Dublin environmental persistence among dairy herds in Pennsylvania
Objective 3: Isolate and screen bacteriophages for potential therapeutic intervention
3.1 Environmental Sampling
During sampling for objective 2, milkhouse wastewater and manure lagoons, where applicable, were sampled by a research technician. Using the manual sampling method outlined by the US EPA18, sterile 50 mL conical vials were dipped into the wastewater source with the mouth facing the current to collect the sample without overfilling. The outside of the sample container was sanitized with 70% ethanol and stored on ice for transport from the farm to laboratory.
3.2 Isolation
Adhering to the methods outlined by Carey-Smith et al.,19 wastewater was diluted 1:10 in SM buffer and incubated at 4°C overnight. Following centrifugation at 1600g for 25 minutes, samples were filtered through a disposable 0.22mM pore-size filter. 100mL of this filtrate was added to a soft agar overlay containing 100mL of a confirmed S. Dublin isolate in the exponential growth phase. Once the overlays were poured and solidified, plates will be incubated at 37°C for 24 hours and then assessed for plaque formation.19 Plaques, if present, will be purified through subsequent serial dilution and plating in soft agar overlays as outlined previously.20
Once purified, phages will be serially diluted in SM buffer. Each dilution will be plated three times and the dilution exhibiting confluent lysis will be chosen for phage recovery and stock creation. To recover phages from the soft-agar layer, 5 mL of SM buffer will be added to each plate and left to incubate at room temperature for a minimum of 60 minutes, swirling the plates regularly. In addition to this liquid, the soft-agar layer will also be removed and added to a conical vial containing 20mL of SM buffer. Once shaken for 30 minutes, the vials will be centrifuged at 1300g for 10 minutes. Once the supernatant is filtered through a disposable 0.22 mM pore-size filter, 0.2% chloroform will be added.19
3.3 In vitro assessment of host specificity
To determine the host specificity of the phages isolated in 3.2, 1:10 dilutions of the phage stock will be plated in soft-agar overlays inoculated with one of the following bacteria: S. Dublin, Salmonella Enteritidis,21 Salmonella Typhimirium,21,22 Salmonella Heidelberg,23 and Escherichia coli. E. coli will act as one of two negative controls. Overlay inoculation with E. coli and the isolated phages should not yield plaques. Additionally, the use of a known E. coli specific bacteriophage in overlays with Salmonella spp. will serve as a phage negative control. An overlay containing both E. coli bacteria and E. coli bacteriophage will confirm the viability of the E. coli bacteriophage and serve as the positive control as noted in the attached figure. These overlays will be created in triplicate for each isolated phage and incubated overnight at 37°C for 24 hours before inspection of plaque formation. Plaque formation in the overlay containing the environmental phage isolates and S. Dublin and the absence of such plaques in overlays containing the same phage isolate but other bacteria inoculum will confirm the host specificity of the isolate phage stock against S. Dublin.
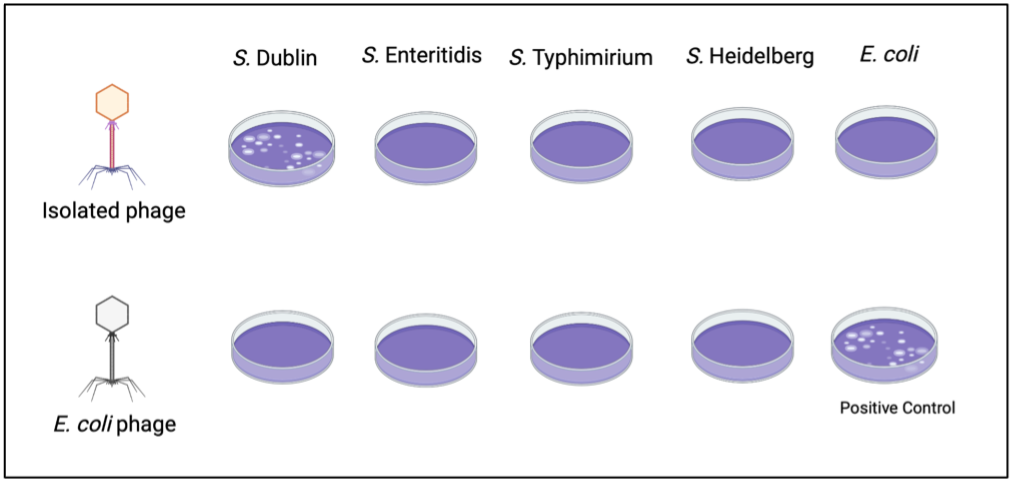
Anticipated Objective Outcomes:
Isolation and confirmed host specificity of a bacteriophage targeted S. Dublin
Citation List
[1] Harvey, R. R., Friedman, C. R., Crim, S. M., Judd, M., Barrett, K. A., Tolar, B., Folster, J. P., Griffin, P. M., & Brown, A. C. (2017). Epidemiology of Salmonella enterica serotype Dublin infections among humans, United States, 1968–2013. Emerging Infectious Diseases, 23(9), 1493–1501. https://doi.org/10.3201/eid2309.170136
[2] Abuelo, A. (2020). Salmonella Dublin in dairy calves. https://www.canr.msu.edu/news/salmonella-dublin-in-dairy-calves
[3] Goodman, L. B., McDonough, P. L., Anderson, R. R., Franklin-Guild, R. J., Ryan, J. R., Perkins, G. A., Thachil, A. J., Glaser, A. L., & Thompson, B. S. (2017). Detection of Salmonella spp. in veterinary samples by combining selective enrichment and real-time PCR. Journal of Veterinary Diagnostic Investigation, 29(6), 844–851. https://doi.org/10.1177/1040638717728315
[4]Cummings, K. J., Virkler, P. D., Wagner, B., Lussier, E. A., & Thompson, B. S. (2018). Herd-level prevalence of Salmonella Dublin among New York dairy farms based on antibody testing of bulk tank milk. Zoonoses and Public Health, 65(8), 1003–1007. https://doi.org/10.1111/zph.12523
[5] Northeastern States’ Ranking in the Nation’s Agriculture: 2018 1. (n.d.).
[6] Mohammed, M., Delappe, N., O’Connor, J., McKeown, P., Garvey, P., & Cormican, M. (2016). Whole genome sequencing provides an unambiguous link between Salmonella Dublin outbreak strain and a historical isolate. Epidemiology and Infection, 144(3), 576–581. https://doi.org/10.1017/S0950268815001636
[7] Paudyal, N., Pan, H., Elbediwi, M., Zhou, X., Peng, X., Li, X., Fang, W., & Yue, M. (2019). Characterization of Salmonella Dublin isolated from bovine and human hosts. BMC Microbiology, 19(1), 226. https://doi.org/10.1186/s12866-019-1598-0
[8] Eyler, A. B., M’ikanatha, N. M., Xiaoli, L., & Dudley, E. G. (2020). Whole‐genome sequencing reveals resistome of highly drug‐resistant retail meat and human Salmonella Dublin. Zoonoses and Public Health, 67(3), 251–262. https://doi.org/10.1111/zph.12680
[9] Lin, D. M., Koskella, B., & Lin, H. C. (2017). Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World Journal of Gastrointestinal Pharmacology and Therapeutics, 8(3), 162. https://doi.org/10.4292/wjgpt.v8.i3.162
[10] Golkar, Z., Bagasra, O., & Gene Pace, D. (2014). Bacteriophage therapy: A potential solution for the antibiotic resistance crisis. In Journal of Infection in Developing Countries (Vol. 8, Issue 2, pp. 129–136). https://doi.org/10.3855/jidc.3573
[11] Squires, R. (2018). Bacteriophage therapy for management of bacterial infections in veterinary practice: what was once old is new again. New Zealand Veterinary Journal, 66(5), 229–235. https://doi.org/10.1080/00480169.2018.1491348
[12] Gordillo Altamirano, F. L., & Barr, J. J. (2019). Phage therapy in the postantibiotic era. In Clinical Microbiology Reviews (Vol. 32, Issue 2). American Society for Microbiology. https://doi.org/10.1128/CMR.00066-18
[13] Gigante, A., & Atterbury, R. J. (2019). Veterinary use of bacteriophage therapy in intensively-reared livestock. In Virology Journal (Vol. 16, Issue 1, p. 155). BioMed Central. https://doi.org/10.1186/s12985-019-1260-3
[14] Nielsen, L. R. (2013). Review of pathogenesis and diagnostic methods of immediate relevance for epidemiology and control of Salmonella Dublin in cattle. In Veterinary Microbiology (Vol. 162, Issue 1, pp. 1–9). Elsevier. https://doi.org/10.1016/j.vetmic.2012.08.003
[15] Baggesen, D. L., Nielsen, L. R., Sørensen, G., Bødker, R., & Ersbøll, A. K. (2007). Growth inhibitory factors in bovine faeces impairs detection of Salmonella Dublin by conventional culture procedure. Journal of Applied Microbiology, 103(3), 650–656. https://doi.org/10.1111/j.1365-2672.2007.03292.x
[16] Davis, M. A., Hancock, D. D., Besser, T. E., Daniels, J. B., Baker, K. N. K., & Call, D. R. (2007). Antimicrobial resistance in Salmonella enterica serovar Dublin isolates from beef and dairy sources. Veterinary Microbiology, 119(2–4), 221–230. https://doi.org/10.1016/j.vetmic.2006.08.028
[17] Wernicki, A., Nowaczek, A., & Urban-Chmiel, R. (2017). Bacteriophage therapy to combat bacterial infections in poultry. Virology Journal, 14(1), 179. https://doi.org/10.1186/s12985-017-0849-7
[18] US-EPA, Region, Sesd, Athens, & Ga. (n.d.). Wastewater Sampling.
[19] Carey-Smith, G. V, Billington, C., Cornelius, A. J., Hudson, J. A., & Heinemann, J. A. (n.d.). Isolation and characterization of bacteriophages infecting Salmonella spp. https://doi.org/10.1111/j.1574-6968.2006.00217.x
[20] Adams, M. H. (1959). Bacteriophages. New York, Interscience Publishers
[21] Singh, V. (2013). Salmonella Serovars and Their Host Specificity. Journal of Veterinary Science & Animal Husbandry, 1(3), 1. https://doi.org/10.15744/2348-9790.1.301
[22] Kingsley, R. A., & Baumler, A. J. (2000). Host adaptation and the emergence of infectious disease: the Salmonella paradigm. Molecular Microbiology, 36(5), 1006–1014. https://doi.org/10.1046/j.1365-2958.2000.01907.x
[23] Hoffmann, M., Zhao, S., Pettengill, J., Luo, Y., Monday, S. R., Abbott, J., Ayers, S. L., Cinar, H. N., Muruvanda, T., Li, C., Allard, M. W., Whichard, J., Meng, J., Brown, E. W., & McDermott, P. F. (2014). Comparative genomic analysis and virulence differences in closely related salmonella enterica serotype heidelberg isolates from humans, retailmeats, and animals. Genome Biology and Evolution, 6(5), 1046–1068. https://doi.org/10.1093/gbe/evu079
[24] Pecoraro, H. L., Thompson, B., & Duhamel, G. E. (2017). Histopathology case definition of naturally acquired Salmonella enterica serovar Dublin infection in young Holstein cattle in the northeastern United States. Journal of Veterinary Diagnostic Investigation, 29(6), 860–864. https://doi.org/10.1177/1040638717712757
Of the 72 different farms enrolled in the study, we identified four S. Dublin positive herds, two of which opted into environmental sampling. Those that opted into sampling were enrolled via the efforts of Penn State University Extension veterinarians. Those that declined were enrolled via the Lancaster DHIA. Environmental sampling was conducted at the two dairies that consented following bulk tank sampling and result disclosure. Swabs taken from maternity pen, group calf housing, and milkhouse locations yielded Salmonella. Of the 23 Salmonella isolates cultured from environmental samples, none were PCR-positive as S. Dublin. As no S. Dublin isolates were able to be successfully cultured from environmental sampling in Objective 2, we proceeded with Objective 3 using a previously serotyped strain of Salmonella Dublin. Despite success in the plaque assay with the E. coli phage control, no phages were successfully isolated from the manure lagoon or milkhouse wastewater samples.
Despite the obstacles to farm enrollment and the limitations this placed on the subsequent study objectives, we were nonetheless able to identify farms in Pennsylvania experiencing issues with S. Dublin, an important starting point to improving awareness and the implementation of risk management strategies. While unable to isolate S. Dublin from environmental samples, we successfully obtained other Salmonella. The lack of S. Dublin specifically in these isolates could be explained by 1) possible enrichment media bias wherein specific media can bias serovar representation after enrichment and/or 2) limitations of colony selection wherein multiple serovars may be present on a streak plate as different colonies and the choice of colony for subsequent culturing and isolation may yield a different result colony-to-colony.
Given the status of Pennsylvania in national dairy production combined with the threat Salmonella Dublin poses to dairy herd health and the sustainability of milk production, an updated understanding of the prevalence of S. Dublin among dairies in the state has been timely. Despite limitations of our project outcomes, the work conducted in the project has been an instrumental first step in more comprehensively addressing this problem. The successful identification of farms experiencing issues with S. Dublin lays the groundwork for further work and establishes the need for spreading awareness of this issue to mitigate dissemination within and between dairy herds in Pennsylvania and surrounding states.
Education & Outreach Activities and Participation Summary
Participation Summary:
A primary short-term goal of this proposal is increased awareness of S. Dublin's significant threat to herd health. This was accomplished through writing an article to be disseminated by Penn State University Extension. This article titled "Prevention and Management of Salmonella Dublin on Dairies" includes information concerning health risk, infection presentation, potential reservoirs, and risk management strategies.
Additional education and outreach activities include an informational talk given at Penn State University Ag Progress Days covering the topics in the extension article, and a second presentation targeted toward clinicians at the 2023 Mid-Atlantic States Conference for Bovine Practitioners. During both of these activities, informational pamphlets were also distributed.
Given our longer-term goal of increasing the sustainability of dairy cattle by improving herd health, our findings will be presented at relevant regional and national conferences such as the 2023 Penn State Dairy Cattle Nutrition Workshop and the American Dairy Science Association annual meeting. The results will also be submitted for peer-review publication in a dairy or veterinary-oriented journal, providing more science-based information for Pennsylvania dairy producers on AMR reduction among their herds. The results of this project will lay out the foundation for a broader effort in quantifying the prevalence of S. Dublin in the Northeastern US, and the application of a multi-pronged approach to mitigate the impact of this pathogen in the dairy industry.
Drs. Hovingh and Springer, who work in collaboration with the USDA-ARS Environmental Microbial and Food Safety Laboratory, hold extension roles and anticipate integrating these results in farm programs related to antimicrobial resistance, antimicrobial stewardship, and promotion of judicious antimicrobial use in calf rearing.
Ongoing activities include the development of three 5-minute “Learn Now Videos”: The first video will explain the particularities of S. Dublin infections in dairy herds and how to spot a potential case. A second video will cover potential management strategies that can be applied by herds to prevent the introduction of S. Dublin into a herd, including biosecurity measures. The third video will focus educating farmers that may be facing issues with S. Dublin on their herd on a previously developed risk assessment tool and how it can be used to improve management and mitigate the effect of S. Dublin infection in positive herds. All videos will be developed by the graduate student in consultation with Penn State Extension personnel (see letters of collaboration) and made publicly available on The Pennsylvania State University extension website to facilitate easy learning.
Combined, these strategies will inform farmers of actions they can be taking to mitigate potential harm to their herds and business as well as inform researchers of the need for continued surveillance and improved strategies for combating antimicrobial resistant pathogens that pose significant threats to livestock health, dairy production sustainability, and food security.
Project Outcomes
Despite limitations of this study, we were nonetheless able to raise awareness of Salmonella Dublin with farmers, researchers, milk testing support staff, and clinicians. Through these conversations over the course of the project, we were informed that our initial contact and discussions had led these individuals to continue the conversation about this issue and the threat it might pose to dairy production in Pennsylvania. Additionally, we were able to identify multiple farms experiencing challenges with S. Dublin and discuss with them possible solutions to the issues they faced. As S. Dublin has the potential to impact the sustainability of dairy in the state via impacts on animal health and milk production, this type of initial awareness is crucial for sharing information about prevention and risk management. In this way, the work conducted through this project combined with our continuation of this research, education, and outreach will contribute to future sustainability by providing researchers, clinicians, and farmers alike with the necessary insights to support dairy production in Pennsylvania in the face of this emerging pathogen.
Throughout this project a great deal of knowledge was gained as it relates to establishing relationships to stakeholders in sustainable agriculture, the practicalities of on farm herd management, limitations and alternatives to culturing Salmonella Dublin specifically as opposed to other serovars. Because of this work, we have established multiple collaborations with other researchers, animal diagnostic labs, veterinarians, and dairy herd improvement associations both within Pennsylvania and throughout the country. The ability to have conversations with a range of stakeholders has improved our understanding of existing perceptions, possible interventions we might propose, and additional strategies to tackling this issue. This project has laid the groundwork for continued investigation of Salmonella Dublin in Pennsylvania, funded by a USDA CARE grant.