Final report for GS17-173
Project Information
Breeding parasite-resistant sheep that are less dependent on the use of anthelmintics to maintain sustainable management and productivity is one of the main goals of Florida Native sheep producers. It would also assist farmers to manage the growing anthelmintic-resistance problem in U.S. Very limited research work has been devoted to selection for resistance to gastrointestinal nematode infections using genetic markers in Florida native sheep populations. Thus, it is proposed that the identification of DNA markers that are indicative of parasite resistance (or susceptibility) could improve selection programs. In order to determine the resistance or susceptibility status of sheep, the objective of this project is to develop genetic markers for parasite resistance in Florida native sheep populations.
1. Identify and validate DNA markers in 100 selected genes (including promoter regions, coding regions and non-coding regions) associated with resistance using targeted sequencing in 153 Florida native sheep, previously characterized with different levels of resistance to natural gastrointestinal nematode infections (extreme individuals: highest fecal egg counts and lowest fecal egg counts).
2. Evaluate the changes in the mRNA structure of the genes with DNA markers associated with parasite resistance.
3. Disseminate to producers the potential benefits of this tool in the selection of breeding animals, using the genetic markers showing benefit in resistance level to gastrointestinal nematodes.
Cooperators
- (Educator and Researcher)
Research
Objective 1
Sheep Population and Phenotypic Data
The research protocol was approved by the University of Florida Institutional Animal Care and Use Committee (Approval number 201810108). All animals used in this study were from a commercial Florida Native sheep farm from Ocala, Florida. The selection flock was established in 1998 from an initial group of 80 rams ranked for H. contortus FEC, with high and low FEC rams mated with approximately 100 foundation dams. Since then, animals were selected for resistance to GIN solely on the basis of H. contortus FEC phenotyping and parentage recording. One hundred and fifty three female and male sheep were tested for gastrointestinal nematodes under natural grazing conditions. Animals were born between December 2017 and February 2018 and naturally exposed to parasites since birth. They were approximately 3 and 5 months old on average and were grouped based on age. At the beginning of the study, initial FEC, FAMACHA score, weight and body condition score (BCS) were measured and animals were drenched with levamisole (18 mg per kg of body weight) the same day of initial screening. Then, animals returned to grazing conditions. Ten days post deworming, FEC reduction was confirmed and FAMACHA score, weight, BCS, and a full hematogram of each sheep were performed. This date was used as baseline (0 days) for future phenotype collection dates. Finally, 28 days post baseline (28 days), the same parasitological and hematological parameters were measured and ADG was determined based on weight and days under study. Identification of gastrointestinal nematode genus was carried out from fresh fecal samples collected directly from the rectum of evaluated sheep as described by Roberts and O’Sullivan (1950) at start of the trial (initial FEC) and during baseline (0 days) and 28 days post baseline (28 days). However, due to low and relatively constant numbers for the other species, only eggs of Haemonchus spp were recorded and analyzed.
The Shapiro-Wilk test was used to test all variables for normality. Box-Cox transformation was performed for all the traits recorded to obtain a normal distribution of values. The R “car” library was used to estimate the power parameter λ and carry out square root transformation for FEC (Estrada-Reyes et al., 2018) and logarithmic transformation (log10) for RBC, HGB, WBC, NEU, LC, MC, BC, and EC. For all the traits, the different time points evaluated during the study (initial screening, 0 days and 28 days) and the difference between twenty eight days post baseline and baseline (28 days - 0 days) were included in the analysis. Spearman phenotypic correlations among all the parameters evaluated were obtained using R software (https://cran.r-project.org/).
Genotyping using Capture Sequencing
Genomic DNA from blood samples collected from the jugular vein using vacutainer tubes with anticoagulant EDTA at day 0 post infection was isolated using DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA) according the manufacturer’s instructions and stored at -20°C. The DNA yield was calculated from a spectrophotometric measurement at 260 nm (NanoDrop‐1000, Thermo Scientific), and the purity was assessed using a ratio 260/280 nm.
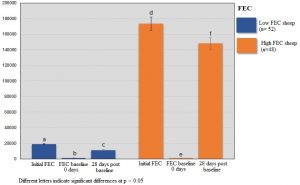
DNA samples (250 ng/μL per sample) from 100 selected sheep samples with the highest (> 1000 eggs per gram of feces, n=52) and lowest (< 900 eggs per gram of feces, n =48) FEC (Supplementary Figure 1) were sent to RAPiD Genomics (Gainesville, FL) for sequencing of 100 genes using DNA Capture sequencing. Briefly, this approach uses a hybrid capture-based enrichment sequencing in which regions of interest are captured by sequence-specific hybridization probes. A focused panel that contained a set of 100 genes was used and custom designed to include genomic regions of interest. Selection of the gene panel was performed based on results from previous studies in sheep (Araujo et al., 2009; MacKinnon et al., 2009; Silva et al., 2012; Venturina et al., 2013; McRae et al., 2014; Benavides et al., 2015, 2016; Sweeney et al., 2016; Berton et al., 2017; Estrada-Reyes et al., 2018). Also, genes related to the immune response against H. contortus and other GIN were considered as candidates for sequencing. The sheep genome assembly from Ovis aries (Oar_v4.0) available at the National Center for Biotechnology Information (NCBI) genome browser was used to design the biotinylated 120-mer probes used for sequencing. Probes were designed to capture sequences at each target gene.
For library preparation, Illumina’s guidelines were followed and Nextera tagmentation, which converts DNA into adapter-tagged libraries, was used. Libraries were denatured and biotin-labeled probes were used for hybridization. Biotinylated probes were captured by streptavidin-coated beads. Then, DNA fragments bound to the streptavidin-coated beads were magnetically pulled down and enriched DNA fragments were eluted. After library preparation, libraries were captured by surface-bound oligos complimentary to library adapters. Then, each library fragment was amplified into distinct, clonal clusters through bridge amplification. After cluster generation, templates were ready for sequencing. Illumina sequencing by synthesis (SBS) chemistry was used for DNA Capture sequencing and samples were then sequenced using a depth of 500 -1000x. Data from sequencing was demultiplexed, cleaned and trimmed. The 3’ ends were trimmed by removing low quality bases with < 20 quality score reads. Clean reads were aligned to genome with MOSAIK software (Lee et al., 2014). For SNP calling, Freebayes software was used and VCFtools (Danecek et al., 2019) was used to generate VCF files. Supplementary Table 1 presents a summary of the genes evaluated in our study.
Quality Control and Association SNP-Based Analysis
Quality control of SNP data set was performed in JMP Genomics 6.0 software (SAS Institute Inc., Cary, NC). A total of 1,546 SNPs were available for analysis. Markers were removed based on genotype call rates per marker (< 95%) and minor allele frequency (MAF) ≤ 0.05, and the final marker dataset consisted of 1,277 SNPs. For the association study, a mixed model was used to evaluate the H. contortus FEC, FAMACHA score, HCT, RBC, HGB, WBC, NEU, LC, MC, BC, and EC. The GenABEL package (Aulchenko et al., 2007) from R software was used to perform the association analysis. Fixed effects included SNP and a contemporary group conformed by age of the animal, sex and group. To control for population structure, the genomic relationship matrix was included in the analysis as random effect and it was calculated from SNP information. The mixed model used for the association analysis was as follows:
y = Xb + Zu + Wa + e
where: y represents the vector of the phenotypic observation for each animal; b is the vector of fixed effects, u is the vector of random animal polygenic effect, u ∼ N (0, Gσu 2), being G the additive genomic relationship matrix; a represents the fixed SNP additive effect, and e represents the residual vector, e ∼ N (0, Iσe2). The matrices X, Z and W represent the incidence matrices for b, u and a, respectively. SNP markers were tested one at a time, and a Bonferroni correction was used to control for multiple testing as follows: alpha = 0.05/number of genes.
Objective 2
Bioinformatics Analysis of Significant SNPs
Information regarding the identity of the genes underlying the significant SNPs associated with different traits was obtained from the sheep genome assembly (Oar_v4.0) available at the National Center for Biotechnology Information (NCBI) genome browser. Then, significant associated SNPs were screened for putative function to evaluate the possible changes in the free energy and the secondary structure of the ovine mRNA. The mRNA sequences from Oar_v4.0 available at the NCBI genome browser were downloaded for the genes with significant SNPs. For each mRNA sequence, a segment of 500 base pairs harboring the SNP in the middle of the sequence was used for prediction of mRNA secondary structures and molecular stability (free energy, G) using the RNA fold web server (Lorenz et at., 2011).
From each association analysis, genes with significant SNPs associated with the evaluated traits were analyzed using Gene Ontology software (Ashburner et al., 2011) to identify molecular function and immunological pathways. These genes were also used in the construction of a gene network related to H. contortus infection. The Cytoscape v3.7.1 software was used for gene network graphics (Shannon et al., 2003).
Objective 3
Dissemination
A small ruminant production symposium will be organized at the Department of Animal Sciences in the University of Florida. Furthermore, abstracts will be developed for meetings such as Annual Meeting of the American Society of Animal Science and National Association of County Agricultural Agents on year two to present the results obtained from this project. Moreover, 2 articles will be developed and published in the following journals: Journal of Animal Science and Shepherd magazine.
Supplementary Table 1 TABLES GWAS FL NATIVE SHEEP 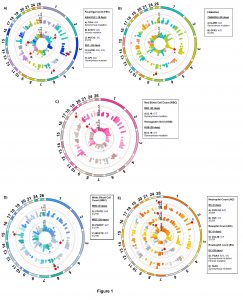
Supplementary Figure 1
Phenotypic Data
The gastrointestinal nematode genus identified from eggs were the following: Haemonchus spp, Moniezia spp, Coccidia spp, and Strongyloides spp. Table 1 shows the descriptive statistics for parasitological and hematological traits available for this study. Table 2 presents the phenotypic correlations between H. contortus FEC and other traits. Significant moderate negative correlations were observed between H. contortus FEC (initial and 28 days) and RBC, and HGB ( < 0.01). For HCT at 28 days, a significant moderate negative correlation was observed with initial H. contortus FEC (p = 0.00005), and with H. contortus FEC at 28 days (p = 0.00009). Significant moderate positive correlated responses were observed between initial H. contortus FEC and WBC (0 days and 28 days), (p = 0.0001 and p = 0.0008, respectively). Also, H. contortus FEC at 28 days was positively correlated with WBC at 0 and 28 days (p = 0.0003 and p = 0.0002, respectively). Finally, significant moderate positive correlations were observed between ADG and H. contortus FEC (initial and 28 days), (p = 0.03 and p = 0.013, respectively). For FAMACHA, BC and EC, no significant correlations were observed with H. contortus FEC (initial and 28 days). A significant strong positive correlation (0.89), (p = 2.2 X 10ˉ16) between HGB at 28 days and RBC at 28 days was observed. Supplementary Table 1 shows the phenotypic correlated responses between all the traits under study.
Genomic Regions identified by the Association Analysis
A total of 23 SNPs covering 14 genes were associated with H. contortus FEC, FAMACHA, RBC, HGB, WBC, NEU, BC, and EC. No significant variants were detected for HCT, LC and MC. A summary of the significant SNPs associated with these traits is presented in Table 3. Overall, associated SNPs identified in this study were located on intergenic and untransated regions (UTR) of 14 genes in OAR1, 2, 3, 4, 6, 7, 11, 15, 18, 19, 20, 24, and 26 (Figure 1). The proportion of variance explained by majority of the significant SNPs associated with FEC, FAMACHA score, WBC, RBC, BC and EC was less than 0.2. Thus, based on the current and previous studies, it is possible that many genes from the immune response (with small effects) are contributing to parasite resistance/susceptibility in Florida Native sheep populations.
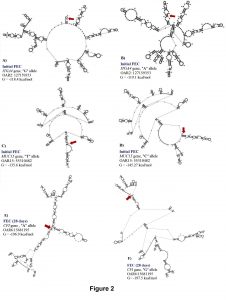
For initial H. contortus FEC, SNPs located in exon 24 of ITGA4 (OAR2:127159353), intron 13 of STAT3 (OAR11:41860577), and the 5’ UTR of MUC15 (OAR15:55310482) genes were significant. These proportion of variance explained by these SNPs was 0.14, 0.10 and 0.11, respectively, for initial H. contortus FEC. The OAR2:127159353 is a G to A substitution which codes for a synonymous codon (Val), where the GG genotype (1,179.4 ± 938.37 eggs/gram of feces) had lower FEC when compared with the AG (3,239.94 ± 2,094.891 eggs/gram of feces) and AA (6,890.1 ± 4,099.9 eggs/gram of feces) genotypes. For this polymorphism, moderate changes in mRNA stability were observed for the “G” allele (-118.4 kcal/mol) when compared with the “A” allele (-119.1 kcal/mol), with great changes in mRNA secondary structure.
The OAR11:41860577 generates a G to C substitution, and OAR15:55310482 confers a base pair change from T to C. For OAR15:55310482, only two genotypes where observed, and the TT genotype (945.6 ± 804.1 eggs/gram of feces) had lower FEC when compared to the CT genotype (4,444.4 ± 2,300.3 eggs/gram of feces). For this SNP, the “T” allele had greater mRNA stability (-135.6 kcal/mol) when compared with the “C” allele (-145.27 kcal/mol) and strong changes in mRNA structure were <spanstyle="font-family:'times new="" roman'"="">observed (Figure 2).</spanstyle="font-family:'times>
Integrin subunit alpha 4 (ITGA4) gene codes for a cell adhesion molecule related to eosinophil mobilization (Rothenberg et al., 2001). Therefore, the protein complex co-formed by ITGA4 protein and beta 1 or beta 7 subunits, regulates cell motility and migration. Thus, it is possible that eosinophils use this protein complex to be mobilized to the area of infection through IL-5 independent or dependent mechanisms. Eosinophils are produced in the bone marrow and are commonly found in almost all the sheep gastrointestinal tract. Sheep eosinophilia is related to reduction in H. contortus larvae establishment (Terefe et al., 2007; Balic et al., 2006). In calves exposed to Ostertagia, Cooperia and Nematodirus spp., higher levels of ITGA4 expression is observed in the mesenteric lymph nodes of resistant animals. In peripheral blood from Chinese indigenous white goats exposed to H. contortus, ITGA4 gene was identified as upregulated in resistant individuals with lower FEC when compared with susceptible animals (Bhuiyan et al., 2017).
The STAT3 gene belongs to the STAT family genes known as signal transducer and activator of transcription. STAT3 protein can be activated by phosphorylation in response to several key activators such as IL-6, IL-10, IL-23, IL-21 and IL-11 cytokines to initiate Th17 signaling and IL17 production. Stronger Th17 response is observed in resistant Caribbean hair sheep at 3 days post-challenge with H. contortus (MacKinnon et al., 2009). Therefore, excretory and secretory proteins from H. contortus larvae are linked to increased Th17 activation and IL17 production (Gadahi et al., 2016). Thus, after initial mechanisms of parasite rejection and complement activation, Th17 response is activated to regulate pathogen clearance and tissue inflammation.
Mucin 15 (MUC15) gene produces a high molecular weight membrane bound glycoprotein consisting of a mucin peptide backbone and O-linked oligosaccharides. Epithelial surfaces of the gastrointestinal tract are composed of these types of proteins and other mucins to protect against gastrointestinal nematodes and represent the first line of defense against pathogens (Millet et al., 1984; Moncada et al., 2003). It is possible that some of these type of mucins are ligands for gastrointestinal nematode adhesins (McGuckin et al., 2011). In Dorper x Red Maasai sheep, MUC15 was identified as potential candidate marker for FEC (Benavides et al., 2015). Thus, ITGA4 and MUC15 could play an important role during the initial establishment and rejection of H. contortus in Florida Native sheep.
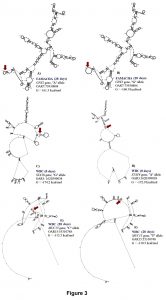
For H. contortus FEC at 28 days and the difference between 28 days and 0 days, the same significant SNPs were identified within the 5’UTR of IL2RB (OAR3:180166632 and OAR3: 180167554) and exon 6 of CFI (OAR6: 15681195) genes. The proportion of variance explained by these polymorphisms was 0.14, 0.13 and 0.15, respectively, for FEC at 28 days. These SNPs are substitutions from G to A, C to G, and G to A, respectively. No significant changes were observed in the stability and secondary structure of the mRNA for OAR3:180166632 in the IL2RB gene. For OAR3: 180167554, the CC genotype (3,451.9 ± 1,725.2 eggs/gram of feces) presented more FEC when compared with the GC (2,397.3 ± 1,793.3 eggs/gram of feces) and GG (677.2 ± 539.3 eggs/gram of feces) genotypes, with no significant changes in stability and secondary structure of the mRNA (Figure 2).
The OAR6: 15681195 is a synonymous mutation with a minor change in mRNA stability of 0.6 kcal/mol but strong impact on the mRNA secondary structure (Figure 2). For this polymorphism, the GG genotype (249.9 ± 124.9 eggs/gram of feces) had lower FEC when compared with the AG (2,370.7 ± 1,775.2 eggs/gram of feces) and AA (1,142.1 ± 909.8 eggs/gram of feces) genotypes.
Complement factor I (CFI) gene encodes a serine protease implicated in regulation of the complement cascade. In hair sheep from the Canary Islands, high expression of CFI gene was observed in resistant animals (Guo et al., 2016). In our study, a genetic variant within this gene was associated with FEC at 28 days suggesting that complement activation is one of the earliest events in the Florida Native sheep immune responses to protect against H. contortus For FAMACHA at 28 days, a significant SNP (OAR3:180148777) in exon 12 of IL2RB associated with this trait. The proportion of variance explained by OAR3:180148777 was 0.13. This SNP (G/A) is part of a synonymous codon and no significant changes in stability and secondary structure of the mRNA were observed. For this locus, the GG genotype (2.11 ± 0.64) had lower FAMACHA score when compared with AG (2.85 ± 0.67) and AA (2.95 ± 0.58) genotypes. Also, for FAMACHA at 28 days, one SNP (OAR7:73930804) in the 3’UTR of GPX2 gene was significantly associated with this trait and explained 0.03 of the proportion of variance observed. The OAR7:73930804 generates an A to G substitution which results in minor changes in mRNA stability (a difference of -0.4 kcal/mol between variants) and mRNA secondary structure (Figure 3).
Similar findings have been observed in Santa Inȇs sheep exposed to natural H. contortus infections, where genetic variants within IL2RB gene are associated to hematocrit (Berton et al., 2017). IL2RB protein is part of the interleukin two receptor complex (IL2R) co-formed by IL2RA, IL2RB and IL2Rγ subunits, and it is activated by IL2 cytokine. This cytokine activates Treg cells, a subpopulation of T cells responsible for maintenance of immunological self-tolerance and homeostasis through immune suppression. Long term infection with gastrointestinal nematodes is linked to activation of Treg populations (Smits and Yazdanbakhsh, 2007). The exact role of Treg response during H. contortus infections in Florida Native sheep remains unclear but a faster switch from Th1 to Th2/Treg response is observed in resistant Suffolk sheep exposed to Teladorsagia circumcincta (Hassan et al., 2011).
The GPX2 gene encodes the glutathione peroxidase 2, which catalyzes the reduction of peroxides and protects against oxidative damage in the epithelium of the gastrointestinal tract. The generation of host oxidants such as reactive oxygen and nitrogen species, is important for parasite control (Ingham et al., 2008; Patel et al., 2009). In Perendale sheep, expression of GPX2 was upregulated in the duodenum of susceptible individuals when compared to resistant animals (Keane et al., 2006). In contrast, expression of GPX2 gene was upregulated in resistant sheep infected with H. contortus (Menzies et al., 2010) during initial days of infection. Similar findings were observed in resistant Australian sheep, where an increase in the expression of GPX2 gene is related to H. contortus challenge during the first days of infection (Lees et al., 2011). Results from these studies suggest that increase of GPX2 expression is essential during larval expulsion in the first days of infection.
For RBC and HGB at 28 days, the same SNP located in IL16 gene (OAR18: 26116868, C/T) was associated with both traits. The proportion of variance explained by OAR18: 26116868 was 0.16 and 0.12 for these traits, respectively. This SNP codes for a synonymous codon (Ser) which did not have significant changes in stability and secondary structure of the mRNA (Figure 3). For OAR18: 26116868 only two genotypes were observed, and the CC genotype presented higher RBC (13.66 ± 1.58 M/μL) and HGB (12.17 ± 1.21 g/dL) when compared with the CT genotype (11.48 ± 1.75 M/μL and 10.57 ± 1.67 g/dL, respectively).
IL16 cytokine is related to inflammatory response and its synthesis is induced by T lymphocytes upon antigen exposure. Also, IL-16 promotes CD4+ T cell production and it is commonly known as lymphocyte chemoattractant factor (O’Shea et al., 2013). In susceptible Perendale sheep selected for high FEC, expression of IL16 gene was downregulated in duodenum tissue (Diez-Tascón et al., 2005). In contrast, in Angus yearlings infected with gastrointestinal parasites, IL16 gene was highly expressed in the mesenteric lymph nodes of susceptible individuals (Araujo et al., 2009).
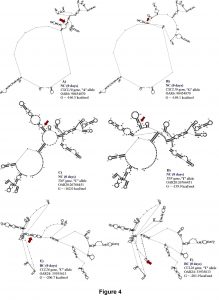
For WBC at 0 days, one SNP (OAR3: 162039038) in the 3’UTR of STAT6 gene was associated with this trait and explained 0.12 of the proportion of variance observed. For OAR3: 162039038, the G to A substitution results in moderate changes in mRNA stability (-1.3 kcal/mol) but great impact on mRNA secondary structure (Figure 4).
Several studies have suggested that mechanisms related to STAT6 activation via IL4 receptor are required for the expulsion of gastrointestinal parasites in sheep (Urban et a., 1998; Venturina et al., 2013). Signal transducer and activator of transcription 6 (STAT6) gene encodes a protein that mediates Th2 signaling. Th2 response is linked to resistance to GIN in sheep (Pernthaner et al., 2005). Commonly features of Th2 response include IL4, IL5 and IL13 cytokine production, eosinophil infiltration and activation mediated by IL5, and IgE production. IgE antibodies bind to antigens expressed in the membrane of gastrointestinal parasites. Then, Fc receptors (FceRI and II) located on the surface of eosinophils recognize IgE antibodies, leading to degranulation and releasing of cytotoxic proteins that damage the parasite membrane (Meeusen et al., 200; McRae et al., 2015). In wool sheep infected with H. contortus, stronger Th2 response is observed at 27 days post-infection, with upregulation of IL4 and downregulation of IFNγRB expression.
Two SNPs (OAR6:49768053 and OAR6:49768057) in the 5’UTR region of PCDH7 gene were associated to WBC at 28 days. The proportion of variance explained by both SNPs was 0.17 and 0.12, respectively. Both polymorphisms generate G to T substitutions with no significant changes in stability and secondary structure of the mRNA. For OAR6:49768053, the GG genotype (10.59 ± 2.34 K/μL) presented higher WBC at 28 days when compared with the GT (10.06 ± 2.17 K/μL) and the TT (6.97 ± 0.40 K/μL) genotypes. For OAR6:49768057, the GT genotype (10.96 ± 2.36 K/μL) presented higher WBC when compared with the GG (10.59± 2.34 K/ μL) and the TT (9.65 ± 2.22 K/μL) genotypes.
Protocadherin 7 (PCDH7) gene encodes a protein from the cadherin family. Cadherins are cell adhesion molecules responsible for cell interactions and maintenance of tissue structure and morphogenesis (Li and Gasbarre, 2010). Studies in Angus yearlings observed upregulation of PCDH7 expression in susceptible animals infected with gastrointestinal nematodes. However, the exact role of PCDH7 protein during H. contortus infections in Florida Native sheep remains unknown.
One SNP (OAR15: 55310748) in the 5’UTR of MUC15 gene was also associated with WBC at 28 days, which results in a base pair change from C to T. The OAR15: 55310748 explained 0.14 of the proportion of variance observed for this trait. This SNP is a putative functional SNP with great changes in mRNA stability (5 kcal/mol) and mRNA secondary structure (Figure 4). For this locus, the CC genotype (10.72 ± 2.50 K/μL) presented higher number of WBC when compared with the CT (10.01 ± 2.09 K/μL) and TT (10.37 ± 2.25 K/μL) genotypes.
For NEU at 0 days, two SNPs (OAR6:90454870 and OAR20:26766451) in the 3’ UTR of CXCL10 and TNF genes were associated with this trait. These SNPs explained 0.14 and 0.13 of the proportion of variance observed. The OAR6:90454870 is an A to G substitution, and OAR20: 26766451 is a C to T substitution. Both polymorphisms are putative functional SNPs with moderate changes in mRNA stability (a difference of 2.6 kcal/mol and -2.1 kcal/mol between variants, respectively) but great impact on mRNA structure. For the OAR6:90454870, the “A” allele presented higher mRNA stability (-166.5 kcal/mol) than the “G” allele (-169.1 kcal/mol), (Figure 5). The AA genotype (2.58 ± 1.62 K/μL) had lower NEU when compared with the AG (3.19 ± 0.68 K/μL) and the GG (6.82 ± 0.72 K/μL) genotypes.
For OAR20:26766451 located on TNF gene, the CC genotype (3.20 ± 1.98 K/μL) had higher NEU when compared to the CT (2.27 ± 0.91 K/ μL) and the TT (1.94 ± 0.61 K/μL) genotypes, with minor changes in mRNA stability but moderate impact on mRNA secondary structure (Figure 5).
The TNF and CXCL10 genes encode proteins related to inflammatory response. TNF protein is commonly secreted by macrophages. CXCL10 protein is produced by monocytes and its production is induced by interferon gamma (IFNγ). In Santa Inȇs sheep, polymorphisms within CXCL10 gene have been associated with platelet count. Thus, it is possible that TNF and CXCL10 genes are related to inflammatory responses in Florida Native sheep exposed to natural H. contortus infections.
For BC at 0 days, one SNP (OAR24:33935613) in the 3’UTR region of CCL26 gene was associated with this trait. This SNP explained 0.16 of the proportion of variance observed. The C to T substitution from OAR24: 33935613 results in a great change in mRNA stability of -4.8 kcal/mol and has a great impact on mRNA secondary structure. The “T” allele (-201.9 kcal/mol) had greater mRNA stability than the “C” allele (-206.7 kcal/mol), (Figure 5). The TT genotype (0.14 ± 0.13 K/μL) presented higher BC when compared to the CT (0.07± 0.06 K/μL) and the CC (0.05 ± 0.04 K/μL) genotypes.
The CCL26 protein is a member of the eotaxin family. Mature eosinophils are activated and migrate to the site of infection in response to various chemoattractants, such as IL-5, CCL11, CCL24 and CCL26 (Rosenberg et al., 2103). Upregulation of CCL26 gene has been observed in draining lymph node tissue from Martinik x Romane back-cross resistant sheep exposed to H. contortus (Sallé et al., 2014). Based on these results, eosinophil migration could play an important role to control H. contortus in Florida Native sheep.
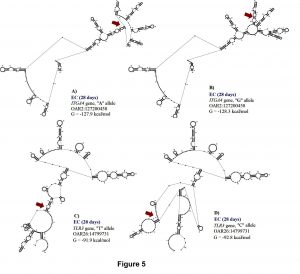
Two SNPs (OAR2:127200458 and OAR26: 14799731) in ITGA4 gene and exon 2 of TLR3, were associated with EC at 28 days. These polymorphisms explained 0.13 and 0.15 of the proportion of variance observed for EC at 28 days. The A to G substitution from OAR2:127199427 results in a minor change of -0.4 kcal/mol in mRNA stability but a moderate change in mRNA secondary structure (Figure 6). Minor changes in mRNA stability were observed for the “A” allele (-127.9 kcal/mol) when compared to the “G” allele (-128.3 kcal/mol). For this SNP, the GG genotype (0.39 ± 0.24 K/μL) had greater EC when compared with GA (0.27 ± 0.23 K/μL) and AA (0.19 ± 0.18 K/μL) genotypes.
The T to C substitution from OAR26: 14799731 results in a minor change of 0.9 kcal/mol but has a strong impact on mRNA secondary structure (Figure 6). The “T” allele (-91.9 kcal/mol) presented greater mRNA stability than the “C” allele (-92.8 kcal/mol), and the TT genotype (0.39 ± 0.37 K/μL) had greater EC than the CT (0.16 ± 0.15 K/μL) and CC (0.24 ± 0.23 K/μL) genotypes.
Our results could suggest that genetic variation within ITGA4 gene could be essential for controlling H. contortus infections in Florida Native sheep, due to its association with FEC and EC. Toll-like receptors (TLR) are commonly expressed in antigen presenting cells and other immune cells. TLR genes are found upregulated in the gut mucosa of resistant sheep infected with H. contortus and Trichostrongylus colubriformis (Ingham et al., 2008). TLR3 signaling could be one of the first immune response mechanisms activated by H. contortus in Florida Native sheep.
Overall, significant SNPs associated with FEC, FAMACHA score, WBC, NEU, BC and EC in Florida Native sheep exposed to natural H. contortus infection are putative functional SNPs that affect the mRNA secondary structure and stability. Some of the genes (ITGA4, STAT3, MUC15, IL2RB and CFI) containing the significant SNPs for FEC were previously highlighted as potential candidate markers for gastrointestinal parasite resistance in sheep populations (Periasamy et al., 2014, Benavides et al., 2015), and identified as differentially expressed in Chinese goats and Angus cattle during H. contortus infection and GIN (Bhuiyan et al., 2017; Araujo et al., 2009).
Results presented in this study confirm that stability of mRNA and conformational changes in the secondary structure of mRNA are important and possible causal mechanisms of gene expression variability. The 5’ UTR region is known to influence mRNA translation efficiency and secondary structures within this region could inhibit translational mechanisms (Leppek et al., 2017). Also, polymorphisms within the UTR regions confer more deleterious effects than polymorphisms in coding regions (Johnson et al., 2011).
Immunological Pathways Related to H. contortus Infection
Five pathways were related to significant SNPs associated with H. contortus FEC. These pathways include JAK-STAT signaling pathway, CD4+ T cell commitment, Th17 signaling, endothelial cell proliferation, and leptin signaling and feeding behavior (Table 4). Leptin is commonly secreted by adipocytes, the placenta, and gastric epithelial cells. High concentrations of leptin in the stomach showed to induce inappetance in rats (Bado et al., 1998). Some studies suggested that increase in leptin concentration is responsible for immune mediated reduction in feed intake in sheep infected with T. colubriformis and T. circumcincta (Stear et al., 2003; Greer et al., 2009). Thus, inappetance could be used by Florida Native sheep as a protective response to minimize feed intake while the damaged mucosa by H. contortus is being repaired.
For FAMACHA, two pathways related to cellular oxidant detoxification and cytokine mediated signaling were identified. For ADG, and RBC/HGB, pathways related to positive regulation of T cell proliferation and cytokine activity were observed. The related pathways to WBC included T helper differentiation and commitment, CD4+ T cell differentiation, IL4- mediated signaling and positive regulation of isotype switching to IgE. For NEU, pathways related to inflammatory response were identified. Finally, for BC and EC, pathways such as CCR3 chemokine receptor
Immune response pathways associated with genes containing significant SNPs for WBC were related to CD4+ T cell differentiation and Th2 response (IL4 signaling and isotype switching to favor IgE antibody production). From these genes, polymorphisms in MUC15 gene were also associated with FEC in our study. Although different SNPs were associated with both traits, the 5’UTR of MUC15 gene seems to play an essential role for H. contortus protection. Thus, initial establishment and rejection of H. contortus could be mediated by MUC15 gene and other molecules in Florida Native sheep.
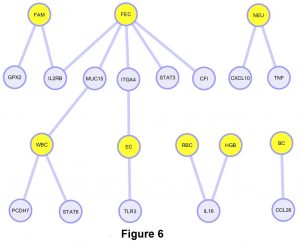
From our gene network diagram, FEC, FAMACHA, WBC and EC had more shared genes with significant SNPs when compared with NEU, HGB, RBC, and BC (Figure 6). Also, FEC was the trait with more associated SNPs in our population. Some of these traits such as FEC and FAMACHA, could be easily incorporated into future breeding programs to control GIN in Florida Native sheep.
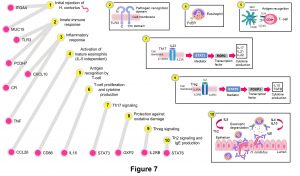
Overall, significant polymorphisms associated with these traits highlighted potential immune response mechanisms related to: initial nematode rejection, innate immune response (inflammatory response and complement activation), activation of eosinophils (IL5 independent), antigen recognition by T-cells, T-cell proliferation and cytokine production, Th17 signaling, protection against oxidative damage, Treg signaling, and Th2 signaling and IgE production. These mechanisms could be representative of the potential immune mechanisms used by Florida Native sheep to control natural H. contortus infections (Figure 7).
Educational & Outreach Activities
Participation Summary:
- Publication of results from this project are published in the Journal of Animal Science: "Association study reveals Th17, Treg and Th2 loci related to resistance to Haemonchus contortus in Florida Native sheep".
- Popular press articles were published in the Shepherd (may 2019 issue p.18) and the EDIS/UF IFAS magazines.
- Conference papers were developed for the 2018 Florida Genetics Symposium and the 17th Annual AMCB Research Symposium from the University of Florida.
- Future presentations will be presented in the small ruminant symposium organized by the University of Florida.
- Extension service was provided to Florida Native sheep producers.
Project Outcomes
Implications
This study is the first report of potential candidate markers for parasite resistance in Florida Native sheep. However, future studies using monospecific infection with H. contortus, genome wide scans and additional animals would be required to validate our findings before these markers could be used with confidence in selection programs. SNPs within 14 genes were significantly associated with FEC, FAMACHA score, WBC, NEU, BC, and EC. These genes were related to JAK-STAT signaling, CD4+ T cell differentiation and commitment, Th17 signaling, leptin signaling and regulation of feeding behavior, cytokine mediated signaling, Th2 response, TLR3 signaling, and CCR3 chemokine receptor binding. It is possible that immune response against natural H. contortus infections is mediated by these pathways in Florida Native sheep. Our results indicate that ITGA4, MUC15, TLR3, PCDH7, CFI, CXCL10, TNF, CCL26, IL16, STAT3, GPX2, IL2RB and STAT6 genes could be potential markers for resistance to H. contortus exposure in Florida Native sheep. Future studies will need to validate these markers in other Florida Native sheep populations before they can be applied.
Economic benefits for farmers
According to to the American Livestock Breeds Conservancy, Florida Native sheep is now on the endangered list of sheep breeds (http://livestockconservancy.org/). To ensure genetic diversity and long-term survival of the breed, protection of the current genetic stock is critical and conservation efforts are required to promote its sustainable breeding and production. The implementation of genomic markers associated to resistance to GIN could improve the selection of Florida Native sheep early in life and benefit the production costs.
As a consequence of the results obtained from this study, sheep farmers increased their sales and started to advertise their flocks as parasite resistant in the Shepherd magazine. Also, farmers started to record the pedigree and performance of animals to facilitate planned mating and breeding. Future research is expected to support sustainable breeding and production of Florida Native sheep producers.
- Provided extension service to Florida Native sheep producers.
- Increased diffusion of genetic selection as a sustainable approach to improve economically important traits in sheep to researchers, students and producers.
It is important to continue supporting small Florida Native sheep producers and local farmers. Florida Native sheep producers are limited source farmers that need support and help from researchers to improve their production. Florida Native sheep breed is endangered and protection of the current genetic stock and conservation efforts are required to promote its sustainable breeding and production. Few research has been devoted to promote sustainable sheep production using genetic selection to improve economically important traits. More public research funds for sheep or goats should be available to improve the american small ruminant industry, support local farmers and transfer cutting edge technology into their systems.