Final report for GS20-224
Project Information
Nitrogen (N) is a main limiting factor for plant growth in agroecosystems. Given the unintended consequences for climate change and environmental impacts caused by highly inefficient N use in modern agroecosystems, increasing nitrogen-use efficiency in agriculture is a central focus of intensive research in agriculture. Mortierella elongata, a member of the early-diverging Mortierellomycota, is a dominant fungus among agricultural soils and functions as saprotroph and root endophyte, affecting several soil processes. Little is known about the effects of M. elongata on soil N transformation processes and the structure of the nitrifying community. Our preliminary results show that M. elongata isolates have a great ability to promote crop growth, but it remains unknown whether the promoting effects of M. elongata on crop growth are associated with its regulation of soil N dynamics and plant-available N. Our recent work shows that integrating two years of bahiagrass (Paspalum notatum) into conventional peanut (Arachis hypogea L.) and cotton (Gossypium hirsutum L.) cropping systems, known as sod-based rotation (SBR), has greater N availability compared with a conventional peanut-cotton-cotton rotation (CR). The proposed work focuses on a plant bioassay using cotton grown in soil from SBR and CR. We will combine stable isotope analysis with molecular tools to quantify the effects of M. elongata on microbially-derived N transformation processes and plant-available N dynamics, as well as on crop productivity under different crop rotations. Our objective is to better predict soil N transformations, helping growers improve economic viability with less external inputs while making agriculture more sustainable.
Objectives
1) Determine the molecular role of Mortierella in regulating soil N transformations and plant N dynamics under different crop rotation systems using 15N tracers;
2) Measure the effects of Mortierella on community structure and functional genes of the soil nitrifying microorganisms under different crop rotations;
3) Quantify the consequences of objectives 1 or/and 2 on cotton productivity.
Hypothesis
1) Mortierella induces higher plant-available N and soil N content and reduces N2O emission under sod-based rotation (SBR) compared to conventional rotation (CR);
2) Mortierella increases the abundance and functional gene activity of the nitrifying community in SBR systems in comparison to CR;
3) The SBR system inoculated with Mortierella will have the highest cotton productivity.
Research
Soil source and bioassay setup for both objectives
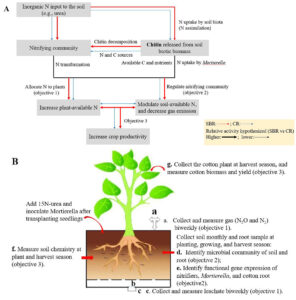
We will set up a plant bioassay comparing CR and SBR soil in a growth chamber at the North Florida Research and Education Center, Quincy, FL. Soils will be collected after cotton harvest in the four-year bahiagrass-bahiagrass-peanut-cotton rotation (SBR) and the three-year peanut-cotton-cotton rotation (CR). Thus, soils will be collected from cotton plots in the single cotton phase of SBR and the two cotton phases of CR (first and second-year cotton). Cotton seedlings be transplanted into closed- system pots (20 cm × 20 cm × 50 cm) (Fig. 1B). M. elongata PMI 624 will be used as the inoculum. Labeled 15N-urea at a rate of 1% (w/w) will be used as N fertilizer to provide 28 kg N ha-1 and trace N transformations. This experiment comprises six treatments (the three cotton soils with or without inoculation with M. elongata PMI 624) sampled destructively at three sampling points, with three replicates per sampling point. This experiment will be assembled in a completely randomized block design, with a total of 54 pots.
Fig. 1 (A) Conceptual view of the central hypothesis that Mortierella increases the efficiency in soil N cycling and crop productivity underlying SBR compared to CR. (B) Mortierella-cotton paired bioassays will be performed to assess our hypotheses.
Objective1. Determine the molecular role of Mortierella in regulating soil and plant N dynamics under different crop rotation systems using 15N tracers
Bulk soil samples will be taken monthly and rhizosphere soil and root will be collected at planting, growing, and harvest season. Gas and leachate will be collected biweekly. The δ15N values (the abundance of stable 15N isotopes) in soils (bulk and rhizosphere soil), gas, microbial biomass, leachate, and root will be measured to determine how M. elongata PMI 624 affects soil N transformations and plant-available N. The isotopic analysis of N2O, N2, soil NH4+ and NO3-, and leachate will be determined by the method of Castellano-Hinojosa et al., (2020), and 15N-enrichment of root will be determined as reported by Andresen et al. (2011). Fluorometric enzyme assays using 4-Methylumbelliferone (MUB) substrates will be used to identify chitinase activity in the bulk and rhizosphere soil and to determine the molecular role of M. elongata PMI 624 on soil and plant N dynamics.
Objective 2. Measure the effects of Mortierella on community structure and functional genes of the soil nitrifying microorganisms under different crop rotations
By analyzing the DNA amplicon sequence data of collected rhizosphere and bulk soil, we can determine the effects of M. elongata PMI 624 on the variation and composition of the soil nitrifying community. After collecting soil and root, samples will be stored at -80 oC. Soil DNA will be extracted using the DNA PowerSoil kit (MoBio, Carlsbad, CA, USA) following the manufacturer’s instructions. Two-step PCR will be used to generate the amplicon library. Bacterial16S rRNA and fungal ITS gene fragments will be amplified using 341F/806R and ITS1/ITS4, respectively. All amplicons will be pooled at equimolar concentrations (20 ng μl-1), and sequenced using Illumina Miseq (v2 250bp, 6Gb sequencing capacity) (Illumina Inc., San Diego, CA, USA). qPCR will be used to quantify the expression of N-associated genes (amoA, nosZ, nirK, and nirS) in the rhizosphere and bulk soil. These results will show how M. elongata PMI 624 affects N transformation processes at the molecular level. Root metatranscriptomics (the analysis of RNA sequence to study gene expression of microbes within natural environments) will be used to measure changes in microbial diversity and N-related functional genes of root with M. elongata PMI 624 inoculation under different cropping systems. Root RNA extraction and cDNA library construction will use the method of Liao et al. (2018). cDNA pools will be sequenced on Illumina HiSeq 2000 instruments in the Duke Center for Genomic and Computational Biology.
The data of bacterial 16S rRNA and fungal ITS gene sequencing will be processed by the QIIME 1.8.0-dev pipeline. RNA sequence assembly and annotation will follow the method of Liao et al. (2018).
Objective 3. Quantify the consequences of objectives 1 or/and 2 on crop productivity
At harvest, we will measure cotton plants for biomass and yields to link results from objectives 1 or/and 2 with crop productivity. Cotton plants will be oven-dried at 65 oC for four days to obtain dry crop biomass. Lint and seed yield will be calculated by grinding a 100g subsample from each pot. Soil properties and chemistry (pH, potassium, calcium, sodium, magnesium, total phosphorus, total organic carbon, and total nitrogen) will be tested to determine the effects of M. elongata PMI 624 on variations in soil chemical properties. Bulk soil samples will be air-dried and sieved to 2 mm, and sieved soil samples will be analyzed for soil chemical properties at the Agricultural and Environmental Services lab of the University of Georgia.
Preliminary data
We evaluated the impacts of three Mortierella isolates (PMI 77, PMI 624, and PMI 93) on plant height, plant dry biomass, and leaf area of different crops using Mortierella-inoculated plants in greenhouse bioassays (Fig. 2). Mortierella isolates had a significant impact on watermelon growth compared to non-inoculated controls. Isolates PMI 624 and PMI 93 significantly promoted at least one variable for corn and tomato relative to PMI 77 and control, and the same effect of PMI 624 and PMI 93 also occurred in the leaf area and plant dry weight of squash. Although all three Mortierella isolates positively affected the growth of bahiagrass, there was a trend of greater bahiagrass growth with inoculation of PMI 624 relative to PMI 77 and PMI 93. However, it remains unclear if the growth-promoting effects of Mortierella isolates are associated with Mortierella-induced N transformations.
Fig. 2 The impacts of Mortierella isolates on the growth of different crop species. Different lower-case letters represent significant differences across different isolates at P < 0.05 by one-way ANOVA (Zhang et al. in review).
- Identifying the ideal substrate and condition for cotton growth
To mitigate the effects of external microbes on cotton performance, we performed surface sterilization of cotton seeds (Deltapine® 1646B2XF and PHY 400 W3FE) before germinating. Specifically, seeds were soaked in distilled de-ionized (DDI) water overnight and then soaked in 10% bleach for 20 min and rinsed with DDI water twice. Afterward, they were soaked in 2% H2O2 for 30 min and rinsed with sterilized DDI water twice. Surface-sterilized cotton seeds were transferred into plastic pots containing autoclaved sand in a growth chamber with 25 °C and 70% humidity and exposed to fluorescent LED lights (200 μmole m−2 s−1) for 12 h per day (Fig. 1A and B). After 14 days of germination (1st true leaf stage), cotton seedlings were transplanted into individual pots containing sterilized sand under the same condition (Fig. 1C).
- Isolating chitin consumers from sod-based rotation (SBR) soils using crab shells
To isolate the native nitrogen (N)-associated fungi as the inocula in this study, we used crab/lobster shell as the chitin substrate (one of the most important organic N sources in the agroecosystem) to culture chitin-consuming fungi (e.g., Mortierella spp.) from sod-based rotation (SBR; bahiagrass-bahiagrass-cotton-peanut) field soil. Specifically, we collected soil from cotton plots under the SBR system. The soil samples were then sieved through a screen with 0.64 cm apertures to achieve a more uniform soil particle size. We sprinkled and embedded flakes of sterilized crab shells in the fresh soil samples and incubated them in petri dishes. After 3, 5, and 7 days of incubation, we pulled some shells and plated very well-washed shells on solid Modified-Melin-Norkrans (MMN) media. Fig. 2 shows the isolated fungal strains.
- Seting up an in-vitro study (culture-based) and applying the RNAseq approach to identify the key genes of representative chitin consumers (e.g., Mortierella sp.) in response to the chitin substrate
To investigate molecular responses of M.elongata (PMI_93) to chitin, we cultured Mortierella in the MMN media with and without the chitin substrate. After 32 hours of culture, we extracted the RNA of PMI_93 fungal mycelia using a CTAB/chloroform extraction, and then the whole cDNA of each sample was sequenced using HiSeq 2000 (Illumina) in the Institute for Genome Sciences and Policy at Duke University. We obtained a total of 157,529,434 (26,254,906 on average) raw reads, and roughly 80% of reads were mapped to the M. elongata genome (M. elongata AG77v2; https://genome.jgi.doe.gov/portal/Morel2/Morel2.download.html). Venn diagram showed that the majority of genes (12,020 genes) were overlapped between Mortierella + chitin and Mortierella – chitin cultures, with only 317 and 312 genes being unique, respectively (Fig. 3A). Further, by performing differential gene expression analysis (cutoff values of |log2FoldChange| >1 and P value < 0.05), we found 239 differentially expressed genes (DEGs) following chitin substrate, with more up-regulated genes (177 genes) compared to down-regulated genes (62 genes; Fig. 3B). This implies that the chitin substrate resulted in a small difference in gene expression. However, principal component analysis of Mortierella genes showed that the expression pattern of these genes treated with the chitin substrate was clearly separated from that of genes without the chitin substrate (Fig. 3C), suggesting that the chitin substrate induced profoundly different expression patterns of whole Mortierella genes.
By annotating DEGs to the EuKaryotic Orthologous Groups (KOG), we found that most up-regulated genes belonged to cellular processes and signaling (31%) and metabolism (31%; Fig. 4A), while over half of down-regulated genes were encoded to metabolism (56%; Fig. 4B). Within these two KOG groups, cell motility, cell wall/membrane/envelope biogenesis, and posttranslational modification, protein turnover, and chaperones were highly expressed in the chitin treatment. As expected, the chitin substrate promoted the expression of genes associated with chitin-related functions, including chitinase, chitin synthase/hyaluronan synthase (glycosyltransferases), and chitinase carbohydrate (Fig. 4C). These suggest that the chitin substrate may promote the growth and activity of Mortierella by providing an important component of cell wall–supporting polymers (e.g., polysaccharides) for the development of fungal cell walls and serving as the C and N source per se. Besides, some genes encoded to chromatin structure and dynamics, RNA processing and modification, transcription, as well as translation, ribosomal structure and biogenesis, were also highly expressed in the chitin treatment, suggesting that the chitin substrate improved the functioning of Moetierella genes involved in genetic information storage and processing. These functions ultimately can promote the development and turnover of Mortierella.
4 Identifying the role of Mortierella elongata in plant performance via a greenhouse bioassay
Mortierella spp. have been demonstrated to have a dual endophyte–saprotroph lifestyle, which may exert plant-growth promotion to many crops via different mechanisms (Zhang et al., 2020). For example, Mortierella may interact with plants and function as synergistic agents, producing antibiotics and phytohormones (e.g., apocarotenoid, indoleacetic acid, and gibberellic acid) (Ozimek et al., 2018; Liao et al., 2019). Mortierella may serve as a saprotroph that drives soil carbon cycling (Clemmensen et al., 2013), P dissolution and immobilization (Detheridge et al., 2016), lipid metabolism, and chitin degradation (Uehling et al., 2017). The Fusarium oxysporum species complex, another major fungal species, comprises a variety of strains ubiquitously living in soils, including plant pathogenic and nonpathogenic strains (Edel-Hermann and Lecomte, 2019). Pathogenic F. oxysporum strains display high functional and genetic diversity (Steinberg et al., 2016), as evidenced by their wide range of plant hosts, including both dicots (e.g., bean, carnation, cotton, and tomato) and monocots (e.g., banana, orchids, and palms) (Edel-Hermann and Lecomte, 2019). However, the pathogenicity of F. oxysporum is host specific. Typically, pathogenic F. oxysporum strains develop two types of symptoms, i.e., vascular wilting that causes Fusarium wilt (a major disease) and root rot (Olivain and Alabouvette, 1999). Fusarium and Mortierella spp. are considered the dominant fungal species in agricultural soils and contribute over 0.5% of the relative abundance in total soil fungal communities. However, how plant-growth-promoting fungi (M. elongata) interact with putative pathogenic fungi (F. oxysporum) and how their interaction affects cotton growth are largely understudied.
To address this question, we performed a greenhouse bioassay by inoculating M. elongate and F. oxysporum in two cotton cultivars (Deltapine® 1646B2XF and PHY400 W3FE). Two months after inoculation, we harvested cotton plants. Harvested plant samples were washed with DDI water twice and wiped by Kimtech Kimwipes Delicate Task Wipers (Kimberly-Clark Corporation, TX, USA). Fresh plant weight and plant height were measured immediately. To avoid biases caused by the different weights and heights among cotton seedlings, we used the changes in plant weight and heights before inoculation with fungi and after harvesting (weight/heightharvest- weight/heightinoculation) to represent plant performance.
There was a significant interaction between fungal treatments and cultivars for changes in cotton biomass (Fig. 5A; P < 0.01). Fungal species only had an impact in the DP1646B2XF cultivar, where F. oxysporum and mixed inocula of M. elongata and F.oxysporum had the lowest cotton biomass change but M. elongata induced the highest cotton biomass change. In contrast, there was no significant difference among fungal treatments in the PHY400 W3FE cultivar, although fungal-inoculated treatments induced a slightly higher weight change as compared to the non-fungal treatment. Similarly, there was a significant interaction of fungal treatment by cultivars for cotton height change (Fig. 5B; P = 0.03). Fungal treatments only affected the height of the DP1646B2XF cultivar, where M. elongata led to the largest height change and mixed inocula had the lowest height change. A cultivar effect was observed in the mixed inocula inoculated treatment, with a significantly larger height change in PHY400 W3FE than DP1646B2XF. Overall, these results suggest M. elongata promoted cotton growth, consistent with our previous study showing that M. elongata exerted plant-growth promotion to many crops (Zhang et al., 2020). We also found that F. oxysporum may function as a pathogen in the DP1646B2XF but not in the PHY400 W3FE, demonstrating that the pathogenicity of F. oxysporum is host specific. The negative effect of the interaction of M. elongate and F. oxysporum on the performance of the DP1646B2XF cultivar suggests that the promoting effect of M. elongata was less than the detrimental effect of F. oxysporum on cotton growth.
- 5. Uncovering molecular mechanisms responsible for the role of chitin in the Mortierella-plant interaction using Metatranscripotimic analysis
To further unravel the effect of chitin addition on the Mortierella-plant interaction, we performed a Mortierella-plant-chitin greenhouse bioassay. Specifically, we inoculated M. elongata in popular seedling roots and then transplanted them into plastic pots that were filled with 180g of sterile play sand. For the chitin treatment, autoclaved crab shells were mixed with sand at a ratio of 2% (w/w). Plants were grown in a growth chamber at 25 °C and 70% humidity and exposed to fluorescent LED lights (200 μmole m−2 s−1) for 12 h per day (Fig. 1A and B). Four months after inoculation, we harvested plant root samples and extracted their total RNA using the CTAB/LiCl extraction method. The whole cDNA of root samples was sequenced using HiSeq 2000 (Illumina) in the Institute for Genome Sciences and Policy at Duke University. We obtained a total of 238,895,950 reads (34,127,993 on average) after filtering low-quality reads, and around 1% of reads were mapped to the M. elongata genome ( M. elongata AG77v2; https://genome.jgi.doe.gov/portal/Morel2/Morel2.download.html). A Venn diagram showed that chitin addition yielded fewer Mortierella genes (8,776 genes) than the no-chitin addition (10,343 genes), with most of the genes (8,166 genes) overlapping (Fig. 6A). Our differential gene expression analysis (with cutoff values of |log2FoldChange| >1 and P value < 0.05) revealed that chitin addition induced more up-regulated Mortierella genes (513 genes) than down-regulated genes (275 genes). This suggests that chitin addition can promote the expression of some genes despite fewer Mortierella genes being detected. A PCA plot showed that there was a clear separation for the expression pattern of all Mortierella genes with and without chitin addition, suggesting that chitin addition can substantially induce Mortierella gene expression in plant roots.
We further filtered highly DEGs with cutoff values of |log2FoldChange| > 1 and P value < 0.01 and annotated these genes using KOG (Fig. 7). Most up-regulated genes were mapped into the group of cellular processes and signaling, metabolism, and information storage and processing (Fig. 7A), while the majority of down-regulated genes were assigned to cellular processes and signaling as well as metabolism (Fig. 7B). Notably, chitin addition induced the high expression of genes associated with cytoskeleton, extracellular structures, nuclear structure, and posttranslational modification, protein turnover, chaperones. This implies that chitin addition promoted the cell development of Mortierella in the plant root. Besides, some highly expressed genes following chitin addition functioned as chromatin structure and dynamics, translation, ribosomal structure and biogenesis, and cell cycle control, cell division, chromosome partitioning, suggesting chitin addition can accelerate cell division and turnover and subsequently facilitate Mortierella growth in plant roots. We also found some highly expressed genes induced by chitin addition were encoded to carbohydrate transport and metabolism (especially three genes associated with chitinase; Fig. 7C), energy production and conversion, and inorganic ion transport and metabolism (including Ca2+ transporting ATPase, Halotolerance protein HAL3 (contains flavoprotein domain), and Na+/K+ ATPase, alpha subunit). N is essential for microorganisms and plants because it is a key constituent of biomolecules including DNA, RNA, chlorophyll, and enzymes (Daly et al., 2021). Our results demonstrate that chitin addition promoted Mortierella growth and turnover in plant roots by fostering the development of cell walls and accelerating cell division, ultimately facilitating the production and transport of N, other nutrients (e.g., K+, and Ca2+), and energy for the growth of microbes and host plants.
6 Disentangling the effect of chitin addition on root-associated fungal communities in the plant-Mortirella system
By aligning high-quality reads to ITS and LSU databases using the MicroFisher workflow, we obtained 31 fungal genera across all samples and then performed microbial analyses at the genus level (Fig. 8). We found that chitin addition decreased the fungal alpha diversity (shown by the Shannon index), although the effect was not significant (Fig. 8A). Principal coordinate analysis (PCoA) and Permanova showed that chitin addition induced a significantly different fungal community composition (P < 0.05; Fig. 8B). These imply chitin addition, an organic N source, had a substantial impact on fungal communities in the plant root, which may play crucial roles in the growth, development, and fitness of host plants (White et al., 2019; Rana et al., 2020). Among 31 fungal genera detected, Neurospora, Fusarium, Mortierella, Peziza, Terfezia, and Linnemannia were the dominant fungal genera (relative abundance > 2%), accounting for over 90% of total reads on average across all samples (Fig. 8C). Not surprisingly, chitin addition significantly increased the relative abundance of Mortierella, consistent with the high expression of Mortierella genes associated with fungal growth and turnover (Fig. 7A). However, chitin addition significantly decreased the relative abundance of Peziza. Peziza is an ectomycorrhizal symbiont of Poplar, which can assist host plants with acquiring nutrients (Tedersoo et al., 2006). This implies plants may develop specific symbiotic strategies for their growth and fitness under different conditions.
Educational & Outreach Activities
Participation Summary:
We published a journal article entitled Mortierella elongata Increases Plant Biomass among Non-Leguminous Crop Species in Agronomy (doi:10.3390/agronomy10050754). In this publication, we found that Mortierella elongata generally promoted metrics of the plant performance among a diverse set of importantly non-leguminous crop species, including the cover crop of SBR (bahiagrass). Our recent EDIS article (The Plant-Growth-Promoting Fungus, Mortierella elongata: Its Biology, Ecological Distribution, and Activities Promoting Plant Growth) provides a brief overview of Mortierella from biological, taxonomical, ecological, and functional perspectives to help readers learn biology and potential modes of action of this fungus. Due to the Covid-19, we had a virtual sod-based rotation field day hosted by North Florida Research and Education Center in October 2020. I worked with my graduate student (Kaile Zhang, co-PI of this student grant project) and delivered a presentation regarding the impacts of the SBR system on soil health-soil C cycling (https://www.youtube.com/watch?v=6a15ROJgXZE&t=304s). In 2021, we led an In-service training (Soil health-Chemical and Biological health) with 22 agents and another 4 professionals (Dr. Ann Blout, Dr. Cheryl Mach, Dr. Yang Lin, and Dr. Sheeja George). This training aims to identify the chemical and biological indicators of different cropping systems across Florida (including SBR). On February 22, 2021, we had virtual training for agents in terms of introducing the importance of soil health in agroecosystems and how to take soil samples from the field. We processed soil samples and analyzing chemical and biological data and generated a potential soil health indicator list using scientific-based evidence. In September 2021, we hosted a round table discussion with the extension agents (15 participants in total) to report the data outcomes. The scope of this IST and round table discussion aim at training the extension faculty to interpret soil biological and chemical data to their producers as the indicators of soil health.
To gain in-depth knowledge on the understanding of roles that soil biota play on soil nutrient cycling under diversified crop rotation systems (in respect to part of the scope in this proposed study), we collected the lectures and led a review article publication (entitled: How soil biota regulate C cycling and soil C pools in diversified crop rotations. Reference ID: 108219). The article has been published in Soil Biology and Biochemistry (doi.org/10.1016/j.soilbio.2021.108219). Besides, our research manuscript studying how long-term SBR affects the nematode community and nematode-microbiome interactions is published in Applied Soil Ecology (doi.org/10.1016/j.apsoil.2021.104254). The research manuscript to study the effects of root microbiomes on cotton performance underlying crop rotation system is published in Biology and Fertility of Soil (doi.org/10.1007/s00374-022-01626-z). Recently, another of our research manuscript submitted to Biology and Fertility of Soil was accepted for publication (entitled "Absolute microbiome profiling highlights the links among microbial stability, soil health, and crop productivity under long-term sod-based rotation"). This work demonstrated how rotation systems shape the structures and stability of soil microbiomes underlying peanut crop. These research publications allow us to investigate the potential interactive effects of rotation, irrigation and environment on the distribution of beneficial microbiome (including Mortierella spp. as the dominant taxa identified).
Project Outcomes
Our project demonstrated that introducing Mortierella elongata into agroecosystems can promote plant growth, with critical implications for sustainable agriculture. Mortierella spp. can act as a saprotroph that drives soil C and nutrient cycling through the decomposition of organic matter. Chitin is a linear polysaccharide of the amino sugar N-acetyl glucosamine, which is an important organic N source in natural ecosystems. Our Mortierella-plant-chitin bioassay showed that chitin addition promoted Mortierella growth and turnover in the plant root by fostering the development of cell walls and accelerating cell division, which may facilitate the production and transport of N, other nutrients (e.g., K+ and Ca2+), and energy for the growth of host plants. This suggests that Mortierella functioned as an endophyte that helped in the biosynthesis of enzymes and bioactive compounds of plants while carrying nutrients from the soil into plants, which could increase nutrient use efficiency.
In modern agroecosystems, N is a main limiting factor for plant growth. However, as it is difficult to predict N fertilizer requirements and many N loss pathways (e.g., N leaching and NH3 volatilization) precisely, N application rates frequently exceed plant requirements. Fertilizer production at the global scale is estimated to equal roughly 100 Tg y-1. Due to low N use efficiency, 50-70% of applied N fertilizer is lost to the environment by leaching of nitrate (NO3-) to aquatic ecosystems and atmospheric losses of NH3 (volatilization) and nitrous oxide (N2O), resulting in unintended consequences for climate change and surrounding environments. Our microbial-based approach of manipulating beneficial microbes together with organic addition would reduce the reliance of agroecosystems on external chemical inputs and mitigate their impacts on ecosystems. Simultaneously, this practice could increase the microbial contribution to nutrient use efficiency, which has the potential to maintain or improve soil health. Overall, this strategy can improve soil quality and agricultural sustainability, increasing crop productivity and decreasing the costs of external inputs, which would help growers increase their net returns.
Making agriculture more sustainable involves managing farmland to remain productive while providing non-provisioning ecosystem services, including greenhouse gas mitigation, efficient nutrient cycling, and economic viability. However, this is challenging in modern agroecosystems. For example, it is difficult to predict N fertilizer requirements and many N loss pathways (e.g., N leaching and NH3 volatilization) precisely. Hence, N application rates frequently exceed plant requirements, leading to unintended consequences for climate change and surrounding environments. With this project, The PI (advisor Liao) and co-PI (Graduate student, Zhang) are proposing that developing microbial-based approaches to increase nutrient use efficiency can increase agricultural sustainability. By performing culture-based studies and Mortierella-plant-chitin greenhouse bioassays, we found that chitin (one of the most important organic N sources in natural ecosystems) can promote the growth and turnover of Mortierella in the plant root by fostering the development of cell walls and accelerating cell division, ultimately facilitating the production and transport of N and other nutrients (e.g., K+ and Ca2+), and energy for the growth of host plants. This is likely a reason why Mortierella can exert plant growth promotion.
Based on the critical role of belowground microbes in C and nutrient cycling, we organized an in-service training (IST) (topic: The importance of soil health in sustainable agriculture) with four additional professionals with different expertise (Dr. Ann Blount, Dr. Cheryl Mackowiak, Dr. Yang Lin, and Dr. Sheeja George). This IST aimed to deliver knowledge to extension agents regarding how different cropping systems shape soil chemical and biological properties to contribute to soil health across Florida. One key task of this IST was to train the agents to identify soil indicators from the farm systems (including biological, physical, and chemical indicators). In February 2021, we had virtual training for 22 agents to introduce the importance of soil health in agroecosystems and how to take soil samples from the field appropriately. After the meeting, trained agents collected farm soil and then sent the samples to us. We (Dr. Liao’s another graduate student, Ms. Erhunmwunse, and co-PI Zhang) led the process and analyses of the chemical and biological properties of these samples. A follow-up meeting was organized to report our results to the agents, where we highlighted the different key biological indicators (including Mortierella) uniquely present in their farming systems. In the report, we also identified which biological indicators (i.e., microbial biomarkers) were sensitive to soil conditions (e.g., pH, N, C), precipitation, and crop management. According to the post-event survey, 85% of the agents were aware of what keystone biological indicators were in their farming systems and gained the knowledge of what potential management strategies for prompting soil health might help to retain these beneficial microbes. To introduce the importance of managing soil microbes in the development of sustainable agriculture to farmers, we attended the 33rd perennial peanut field day in UF/IFAS North Florida Research and Education Center, Quincy, and delivered a poster presentation (entitled” How do mycorrhizal fungi improve nitrogen use efficiency in the rhizome peanut-bahiagrass system”).
We are also developing collaborations with other teams to develop microbial-based strategies to improve agroecosystem sustainability. For example, we are working with Dr. small and Rebecca Barocco (his graduate student) to design a field experiment by implementing multiple cover crops to minimize pest and disease pressure for peanuts. We are also collaborating with Dr. Shahid and his graduate student in verifying whether Mortierella spp. promote the growth of citrus.
Overall, this project advanced our understanding that developing microbial-based strategies is an efficient and effective approach to improving agroecosystem sustainability.
In future studies, we would focus on more complex microbial systems to study their microbe-microbe and microbe-plant interactions in plant growth. Besides, we would apply agricultural soils to perform greenhouse studies, making our studies more representative of modern agricultural systems
Information Products
- Long-term sod-based rotation promotes beneficial root microbiomes and increases crop productivity
- Integrating perennial bahiagrass into the conventional rotation of cotton and peanut enhances interactions between microbial and nematode communities
- How biota regulates C cycling underlying crop rotations. Soil Biology and Biochemistry
- Absolute microbiome profiling highlights the links among microbial stability, soil health, and crop productivity under long‑term sod‑based rotation