Progress report for GS22-268
Project Information
Rhizoma peanut (RP) [Arachis glabrata Benth.] is incorporated into grassland pastures, such as bahiagrass (BG) [Paspalum notatum Flüggé], to increase pasture feed quality and reduce the need for nitrogen (N) fertilizer inputs. Apart from N, phosphorus (P) is important for forage and animal nutrition. The nutrient dynamics of P is of interest because a large percent of soil P is unavailable to plants, especially in weathered soils, and P play a critical role in the contamination of water bodies. This study investigated P uptake and mobilization strategies in BG monoculture versus BG-RP mixture under low and adequate soil P level. We also identified soil microbial communities responsible for nutrient uptake both in the field and in the greenhouse. We hypothesized that in addition to soil and plant root associated microbes, RP will employ strategies that will increase soil P availability in BG-RP mixture, especially under low soil-P. Our findings showed that under low soil P, RP in BG-RP mixture has the potential to increase BG shoot biomass compared to BG monoculture. Another interesting finding was that under low soil P, RP in BG-RP mixture had greater shoot concentration of P, Fe, Mn, Zn, and Cu concentrations than BG shoots under low P, indicating that RP has higher ability to mobilize these nutrients than BG. Soil-P fractions analysis showed that NaOH- organic P occupied a larger percent (34%) of soil P. Soil microbial analysis conducted on the field showed that RP enhanced the relative abundances of fungal genera Fusarium, Gibberella, and Humicola. These fungi contribute to litter decomposition and nutrient cycling through their saprophytic lifestyle.
Hypothesis
- The integration of RP into BG system will enhance the community and functionality of both BG and RP associated microbial groups that are responsible for P solubilization and mineralization in microbial-soil interface and P exchange in plant-microbial interface.
- The enhancement of microbial mediated P cycling in hypothesis 1 will be controlled by a set of P cycling genes (Table 1), including but not limited to the genes responsible for solubilizing inorganic P (e.g., gcd and PqqABCDEF), the genes responsible for mineralizing P (e.g. PhoADN, aphA, olpA, php, glp, and app, and the genes involved in microbial-P exchange and plant uptake at the microbial-root P interface.
- Soil microbial P cycling processes (genes and enzymes release), especially the genes involved in P uptake and transport as well as P starvation genes, will be suppressed when P mineral fertilizers are applied.
Objectives
- Determine the effect of P fertilizer application on soil P pools, plant P uptake and P transfer under BG monoculture versus a BG-RP mixture.
- Identify microbial communities in the different belowground plant parts of bahiagrass (root+rhizome), rhizoma peanut (roots+ rhizome+ nodules), and in the rhizosphere, in response to forage species composition with or without mineral P fertilizer.
- Use qPCR to quantify organic P mineralization (e.g., phoC and bpp) and inorganic P dissolution (e.g., pqqC) genes in rhizosphere soil and bulk soil across forage species with and without P fertilizer.
- Profile genome expression of root microbiomes (including AMF) and root cells, with emphasis on identifying and quantifying the expression of key genes that are involved in P uptake and transport, as well as P-exchange (and P-metabolism) in the different forage systems (monoculture vs mixture) with or without mineral P fertilizer.
- Organize in-person visits with producers in North, Central, and South Florida and present our project findings to them.
Cooperators
Research
Greenhouse experiment setup: Two phases of greenhouse experiment were established to determine the microbial strategies employed by bahiagrass and rhizoma peanut for P and nutrient mobilization and uptake.
First phase: A 3x2 factorial greenhouse experiment with three forage treatments and two P levels, was conducted at the University of Florida, North Florida Research and Education Center (NFREC), Quincy, Florida. Forage treatments included bahiagrass monoculture (BG), bahiagrass + “Ecoturf” rhizoma peanut (BG-RP), and bahiagrass + “Ecoturf” rhizoma peanut separated with a 30 µm fabric (BG-RPF) to prevent physical interactions of roots between both plants. The two P levels were treatments with no added P fertilizer (low P) and treatments with 100 kg P2O5 ha-1 (high P). The treatments were arranged in randomized complete block design with 4 replicates.
Harvest and nutrient analysis for whole plant tissues and soil samples (Objective 1).
Above and belowground biomass were measured. Root and rhizomes (~2 g) were collected within 2 mins and placed in liquid N2 for DNA and RNA analysis. Root subsamples (~1.5 g) were collected for arbuscular mycorrhizal (AM) colonization assessment. The soil and aboveground samples were sent to Soil, Plant, and Water Laboratory, University of Georgia for carbon, nitrogen, phosphorus, and micronutrient analysis. Nutrient uptake for aboveground samples was also measured. Soil samples were analyzed for soil pH, total carbon (TC), total nitrogen (TN), and Mehlich-3 extractable P, K, Mg, Fe, Mn, Zn, and Cu. Soil P fractions (inorganic and organic soluble and insoluble P) were determined using the Hedley Fractionation and data were analyzed using ANOVA in R software.
Identify microbial communities and key genes involved in P transformation in bulk soil, rhizosphere, and root compartments (Objectives 2, 3, and 4).
The DNA from the soil samples were extracted using DNeasy PowerSoil extraction kit, while DNA from root compartments will be extracted using the cetyl trimethylammonium bromide buffer approach. Two-step PCR will be carried out to produce the amplicon library, using the bacterial 16s rRNA primers (341F/806R) and ITS1F/ITS4 and NS31/AML2 for the fungal and AMF communities. The amplicon pools will be sent to Center for Genomic and Computational Biology, Duke University using Illumina MiSeq sequencer (v3 300 PE). The roots from each pot will be stained and viewed under the microscope to count the AMF and their structures.
We will employ metatranscriptomics for the root samples collected. Root RNA extraction and cDNA construction using poly-enrich (for eukaryotic RNA) and Ribo-0 kit (for bacterial RNA) will be carried out modified according to Liao et al. (2014). The cDNA pools will be sent to Duke Center for Genomic and Computational Biology for sequencing using the Illumina Nova seq6000 (150bp-PE, 6Gb per sample) (Illumina Inc., San Diego, CA, USA). The computational workflow will be developed according to Liao et al. (2014) to sort the genes into bacterial and fungal rRNA, and functional genes of the root microbiome respectively and downstream analysis will be carried out in R software.
Second phase: This greenhouse was recently established. A rhizobox was designed to study in detail the microbial mechanisms involved in P and nutrient mobilization and uptake in BG-RP mixture. The treatment design was a 2x2x2 factorial design including BG-RP partitioned by 25 µm fabric and BG-RP without fabric, with or without AMF application, and two P levels were treatments with no added P fertilizer (low P) and treatments with 100 kg P2O5 ha-1 (high P). The treatments were arranged in randomized complete block design with 4 replicates. Experiment is still ongoing, and results collected from this study will be part of our next report.
Field experiment: Three on-farm studies were conducted across Florida (North, Central, and South) as well as two other field studies at NFREC Quincy and Marianna to determine the soil microbial communities associated with integrating rhizoma peanut into bahiagrass pastures. The roles of these microbial communities in nutrient cycling in these pastures were also investigated.
Results from the greenhouse experiment: Shoot biomass measured, and nutrient (C, N, P, Fe, Mn, Zn, and Cu) analyzed from phase 1 greenhouse experiment showed that bahiagrass grown in bahiagrass-RP mixtures may be favored by the presence of RP at low soil P. This finding was only observed at the first harvest (Fig. 1A) and not at the second harvest (Fig. 1B). When we looked at the shoot nutrient concentration of the forages under low and high soil P level, we observed that regardless of P levels, RP had greater N concentration, whereas bahiagrass had greater shoot C concentration. The effect of forage treatment on shoot P concentration was only observed at low P, where RP had greater shoot P than bahiagrass (Table 1). Shoot micronutrients usually associated with P in soil and other micronutrients solubilized by organic acids and other compounds released by plant roots were mainly greater in RP shoots than BG under low P and were similar between both forages under high P. This provides evidence that RP might have superior strategies in mobilizing and taking up these nutrients.
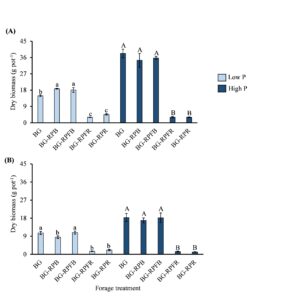
Table 1: Shoot nutrient concentrations of forage treatments at low and high P levels.
|
FT |
C |
N |
P |
Fe |
Mn |
Zn |
Cu |
|
|
-----------g kg-1---------------- |
-------------------mg kg-1---------------------- |
|||||
|
Low P |
|||||||
|
BG |
a467±3.1a |
22±0.4b |
0.6±0.03b |
47±3.3c |
876±36.4b |
57±16b |
9.6±0.5b |
|
BG-RP |
463±1.8a |
22±0.6b |
0.6±0.03b |
41±4.6c |
924±66b |
48±4b |
10.0±1.0b |
|
BG-RPF |
465±4.0a |
21±1.6b |
0.6±0.02b |
45±4.4c |
944±110b |
44±2b |
8.9±0.7b |
|
RP-BG |
403±2.7b |
27±0.8a |
1.7±0.20a |
82±25.1b |
1865±330a |
196±37a |
21.5±2.3a |
|
RP-BGF |
405±1.1b |
27±0.3a |
1.7±0.28a |
147±15.8a |
1857±444a |
231±34a |
24.6±4.2a |
|
High P |
|||||||
|
BG |
464±0.5A |
22±1.2B |
1.4±0.14A |
35±4.3A |
1008±67A |
79±24A |
9.1±1.2A |
|
BG-RP |
452±8.2A |
22±3.0B |
1.6±0.25A |
37±4.4A |
942±55A |
60±15A |
8.0±0.7A |
|
BG-RPF |
454±3.8A |
21±1.3B |
1.6±0.09A |
33±4.0A |
921±113A |
51±11A |
7.7±0.8A |
|
RP-BG |
398±3.7B |
25±2.0A |
1.5±0.22A |
40±6.3A |
414±125B |
72±39A |
10.1±1.2A |
|
RP-BGF |
402±4.0B |
24±2.4A |
1.2±0.29A |
39±8.5A |
566±104B |
94±12A |
8.1±2.4A |
|
|
bP values |
||||||
|
FT |
<.0001*** |
<.0001*** |
0.015* |
0.002** |
0.820 |
<.0001*** |
<.0001*** |
|
P |
0.005** |
0.227 |
0.003** |
<.0001*** |
<.0001*** |
0.005** |
<.0001*** |
|
FT x P |
0.678 |
0.428 |
<.0001*** |
0.009** |
<.0001*** |
0.001*** |
<.0001*** |
aValues are mean ± standard errors. bP values from ANOVA table showing the effect of forage treatment, P fertilizer level, and their interactions on shoot nutrient concentrations. *, **, and *** indicate significance at P< 0.05, 0.01, and<.0001, respectively. Pairwise comparison was carried out using Tukey’s analysis. Lowercase and uppercase letters show significant differences among forage treatments at low and high P levels, respectively.
Results from field experiment:
On-farm studies
The effect of incorporating RP into bahiagrass pastures in Florida on soil microbial communities responsible for nutrient cycling, especially soil bacteria depend on pasture location and growing season. Increased soil bacterial diversity in bahiagrass-RP mixtures was only evident in North Florida and during May sampling. However, the presence of RP consistently enhanced soil fungal diversity across Florida pastures during peak forage production in July (Erhunmwunse et al., 2023).
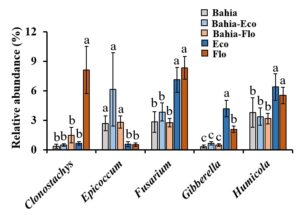
From the field experiment conducted at NFREC, Marianna, the presence of RP in bahiagrass-RP pastures enhanced the relative abundance of dominant soil fungal taxa, such as Fusarium, Gibberella, and Humicola that contribute to soil organic matter decomposition (Figure 2).
References
Erhunmwunse, A.S., Queiroz, L.M.D., Zhang, K. et al. Changes in soil microbial diversity and community composition across bahiagrass and rhizoma peanut pastures. Biol Fertil Soils 59, 285–300 (2023). https://doi.org/10.1007/s00374-023-01701-z
Educational & Outreach Activities
Participation Summary:
We have had opportunities to showcase our project in the greenhouse as well as the field experiment during field days, student tours, in-service training to students, farmers and agricultural professionals. Below are the details of the outreach activities that we have been involved in.
In-service training:
We educated state and county extension agents about the need for improving phosphorus use efficiency in Florida soils using legumes and soil microbes at North Florida Beef and Forage In-service training (FBF-IST) for Florida state and county extension faculty (03/30/2023). Topic: Soil-plant interactions, Role of legumes and microbes in phosphorus mobilization in highly weathered soils, and mycotoxins in forages.
Field day:
We presented a poster (legume cultivars effect on soil microbial communities (the hidden engineers) in bahiagrass system) on the 33rd perennial peanut field day, NFREC, Quincy, FL.
Tours:
- Student tour. 2022. Students in the Department of Soil Water, and Ecosystem Sciences from Gainesville and research centers in Florida. (~ 15 students were in attendance)
- Plant Sciences Council, a graduate student organization tour (~ 12 students were in attendance)
Webinars, talks and presentations:
- Presentation at “STEM in the art” exhibition (11/29/2022) in Tallahassee. Scope: We demonstrated the benefits of perennial peanut as a forage and ornamental purpose in Florida. We taught the audience the importance of N fixing bacteria and other important microbes residing in rhizoma peanut nodules and practiced the art of culturing non-pathogenic bacteria on agar plates. Instructors: Adesuwa Erhunmwunse and Ben Reimer (PhD students). Target audience: Tallahassee community residents.
-
Demonstrations at Tallahassee Science Festival (10/22/2022). Scope: Presented the role of soil microbes (mycorrhiza and rhizobia) in P cycling in agriculture. We shared handouts, used posters, and samples of rhizoma peanut as demonstration materials. Instructors: Adesuwa Erhunmwunse and Ben Reimer (PhD students), Kaile Zhang and Haihua Wang (post docs), and Vijay Verma (Biologist Scientist).
Other Educational Activities
Adesuwa Erhunmwunse (PhD student) trained a master’s student (Rachel Balster) this year on how to set up an experiment and explore molecular and microbial techniques to identify bacterial communities from the nodules of rhizoma peanut.
Project Outcomes
The outcome from this study will provide insights on forage management practices that can increase P use efficiency in Ultisols, reduce the cost of P fertilizer inputs, mitigate environmental degradation, increase pasture productivity, and identify potential microbes that can increase P mobilization and increase nutrient use efficiency in pastures. We promoted our research to the local farming community on the Perennial Peanut Field Day and farmers are eager to see our research findings. They are curious to know if rhizoma peanut grown with grasses will improve P uptake and reduce P fertilizer applied to their pastures. We plan to publish extension articles to communicate our research findings to them. We exchanged contacts with some of the producers to inform them when our article is released. By improving the P fertilizer use efficiency of pasture systems, fertilizer savings will impact all regional forage and livestock producers.
Information Products
- Changes in soil microbial diversity and community composition across bahiagrass and rhizome peanut pastures
- Soil microbes in bahiagrass and rhizoma peanut pastures
- Short-term integration of perennial peanut into bahiagrass system influence on soil microbial-mediated nitrogen cycling activities and microbial co-occurrence networks
- Soil bacterial diversity responds to long-term establishment of perennial legumes in warm-season grassland at two soil depths