Final report for LNC18-405
Project Information
Legumes such as field peas have emerged as a viable option for farmers to diversify their cropping rotations. An added benefit of legumes in rotation is their ability to fix nitrogen through a symbiotic relationship with rhizobia bacteria. The nitrogen fixed by this interaction not only helps the legume crop, but also the subsequent crop as the fixed nitrogen is released to the soil as legumes decompose at the end of the season. This process allows farmers to be less reliant on expensive nitrogen fertilizers for both crops. However, as field peas continue to expand into the northern Plains, where dry, hot summers are common, it is evident that heat and drought stress are major limiting factors to both production and adoption by farmers. The problem of drought stress is two-fold in that it not only affects yield, but it also severely limits nitrogen fixation by rhizobia.
It is possible that native rhizobia, acclimated to the local climate and soils, are more effective at nitrogen fixation under stress conditions. This project intends to 1) examine the paired use of drought tolerant field peas and native rhizobia versus commercial rhizobia to maintain nitrogen fixation in field peas during drought stress, 2) identify the most effective inoculation methods for nitrogen fixation, and 3) create an open-source culture method that farmers can utilize on their own farms to be used as either a standalone inoculant or in combination with commercial inoculant.
Project Outcomes
1) Evaluate the use of native rhizobia to maintain nitrogen fixation in field peas during drought stress and improve crop performance
2) Identify the most drought tolerant FP varieties and the most effective inoculation methods for nitrogen fixation in field peas and ensure that farmers are employing these techniques
3) Educate farmers on the value of effective nitrogen fixation and the importance of legumes in diversified cropping rotations
4) Create an open-source culture method that farmers can utilize on their own farms to be used as either a standalone inoculant or in combination with commercial inoculants
Legumes such as field peas (FP) have emerged as a viable option for farmers to diversify rotations. In fact, SARE
has recently funded several projects examining the value of FP in intensified farming rotations, which replaces the
traditional wheat-fallow system. An added benefit of legumes is their ability to fix atmospheric nitrogen through
symbiotic relationships with ‘rhizobia’ bacteria. This fixed nitrogen helps the legume crop, and also the
subsequent crop as the nitrogen is released to the soil during plant decomposition. This process allows farmers to
be less reliant on expensive nitrogen fertilizers for both crops. To our knowledge, SARE has not funded similar
projects or projects related to rhizobia development.
However, as FP acreage expands across the northern Great Plains, where dry, hot summers are common, it is
evident that heat and drought stress are major limiting factors to production and adoption by farmers. The
problem of drought stress in FP is two-fold. It not only affects yield, but also limits nitrogen fixation by rhizobia.
Drought stress leads to a reduction in nodulation due to limited root hair growth which consequently reduces the
amount of N-fixation (Bordeleau, 1994). Studies have shown that even moderate drought conditions can lead to a
50% or greater reduction in N-fixation (Kirda, 1989). Consequently, farmers lose money on poor yields and lose
the soil health benefits of nitrogen fixation.
We currently conduct FP variety trials at five locations across the state. The past two growing seasons have been
defined by drought and this project stems from our experiences with these variety trials and producer concerns.
There has been some concern that inoculants are failing under drought stress, hence the need for more drought
tolerance in both plant and inoculant. Moreover, during our field tours, farmers remarked that if drought is severe,
a modestly productive legume would still be beneficial as a soil health builder even when not producing a viable
yield. This would be preferable to other crops such as failed wheat or corn. In other words, where a harvest is not
viable due to drought, field peas are a preferable option to wheat due to the low C:N, which degrades rapidly and
provides a valuable green manure.
Native rhizobia populations arise in the soil by two means, A) from compatible strains nodulating wild legumes or
B) commercial strains remaining in the soil from previous cropping seasons (Vessey and Chemining'wa, 2006).
The failure of commercial inoculants under drought conditions is not surprising. It is well established that when
microorganisms are introduced into a new environment they often fail to survive because the environment is too
different from their native growing conditions or they cannot compete well with the native community and thus fail
to become established (Mendes, 2013). This is particularly true for rhizobia where it is recognized that inoculation
efficacy can be improved by using strains that were isolated from environments similar to where they will be used
(Lupwayi, 2006). In spite of this, currently, most commercial rhizobia inoculants are sold as 'one size fits all.' The
same rhizobia strains used in South Dakota are also used in South Carolina.
We envision an integrated approach that matches drought tolerant FP varieties with compatible, drought tolerant
rhizobia adapted to local growing conditions so both nitrogen fixation and yield can be maintained during drought
and heat stress. In order to improve rhizobia survival and activity, we will evaluate the interaction between
commercial and native rhizobia to identify suitable crop variety-rhizobial strain pairs for improved performance
under drought and heat stress. A combination of commercial and native rhizobia may work in concert.
Commercial rhizobia could maximize nitrogen fixation during optimal conditions while native rhizobia will
supplement nitrogen fixation during stress conditions. While the examination of native rhizobia is not a new
concept, evaluating native and commercial inoculants together is. Typically, research on native rhizobia focuses
on the value of the native rhizobia to obviate the necessity of applying commercial inoculants (e.g. Chemining'wa
and Vessey, 2006). But most studies fail to culture the native rhizobia and apply it at similar rates that a
commerical inoculant is applied at. Therefore, while the native population may be effective at nodulating the target
legume, it is not present in sufficient quantities to compete with a commercial inoculant. Moreover, there is little if
any information in the literature that examines how different rhizobial strains might work symbiotically under
varying abiotic stressors.
The hypotheses for this research are: 1) native rhizobia are better adapted to their local environment and can
maintain nitrogen fixation more effectively than commercial rhizobia under stress conditions when supplied in
adequate concentrations. And 2) specific legume variety-rhizobial strain combinations will perform differently due
to compatibility and competitiveness.
Cooperators
Research
Native rhizobia cultured from drought-prone regions can outcompete commercial inoculants under drought conditions in field pea production.
In 2020, soil was gathered from 24 unique sites across Montana, North Dakota and South Dakota. The sites represent different (never cultivated) biomes including high-altitude, mountainous regions, forest, undisturbed prairie and shrub-steppe grassland. Uninoculated field peas were grown in each of these soils under lab conditions and harvested after 8 weeks to determine colonization by native rhizobia. Nearly all soils produced nodules. Of these nodules, 92 rhizobia samples were successfully cultured. These samples will then be screened for drought tolerance and candidate rhizobia will be tested for further greenhouse and field studies in 2020. This process was repeated during the 2021 calendar year.
Lab-Based work:
From the grown field peas that were successfully nodulated from the trap experiments, five representative nodules from plants of each sample site replicate were crushed in Vernon’s Rich (VR) media (For the first 10 glycerol stocks, however, single representative nodules of three listed sites were first used). After being strained through a 70micro meter nylon strainer, 750 microliters of filtrate was mixed with equal parts 50% glycerol solution and frozen in a -80 degree Celsius freezer. This created 91 total glycerol stocks of rhizobia “soup” cultures.
Bacteria colony isolates were acquired by culturing the glycerol stocks in 10ml of liquid VR medium in 50ml glass vials and placed in a shaker (28 degrees at 245 rpm). After successful growth, broth was streaked on 1.2% VR agar plates. Five representative colonies were picked and transferred into Nitrogen-Free media. After 1 week of growth, successful isolates were harvested and stored into glycerol stocks.
We are attempting to determine drought tolerance by both withholding sufficient moisture, but also through the addition of salinity which creates a similar stress on the rhizobia. To determine saline tolerance, isolates were screened in NaCl supplemented liquid VR cultures (3 replicates) and measured at OD600 after a 3-day period. USDA 110, a highly characterized rhizobia, was used as a control to determine reliability of experiment. Below are the results of the initial run (concentrations of 0.0017M, 0.3M, and 0.5M).
Field-based work:
Three native rhizobia species were selected to be field tested along with a commercial and liquid inoculant in 2020. Two varieties of field peas (Carver and Profit) were inoculated with each rhizobia species and planted in 5x30ft plots with 6 replications. Sub-plots were harvested at 50% flowering for measurement of N fixation - determined through the 15N natural abundance method.
In 2021 and 2022, further lab and field testing was conducted. At this point in the research we narrowed our rhizobia search down to just a few promising candidates. These candidates were selected based on previous lab and field-based measurements. The field trials took place at two locations in western and central South Dakota. The stimulate drought conditions, we utilized two planting dates at each location: the first date was considered a normal 'on-time' planting and the second was a late planting, which pushes the critical anthesis period into the warmer part, drier part of the summer growing season.
To date, we do not have conclusive results. Currently, we have 91 rhizobia candidates. It is unclear how many of these samples are unique. Genetic sequencing will be utilized to determine how many species we actually have. However, we have begun the screening process, which involved developing a workflow and a series of criteria from which to evaluate each candidate species. Below are the representative results of rhizobia growth at varying levels of salinity.
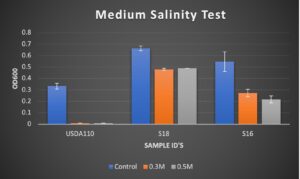
Figure 1. Results of OD600 measurements of bacterial isolates USDA 110, S18, and S16 after 3 days of growth. Treatments used were 0.0017M NaCl (Control), 0.3 M NaCl, and 0.5M NaCl.
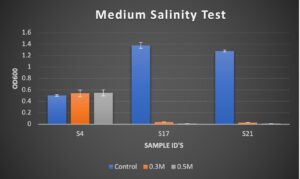
Figure 2. Results of OD600 measurements of bacterial isolates USDA S4, S17, and S21 after 3 days of growth. Treatments used were 0.0017M NaCl (Control), 0.3 M NaCl, and 0.5M NaCl.
Due to having little difference in the ranges of values between 0.3M and 0.5M NaCl, the “Medium” scale, we adjusted experiment to use “High” salinity treatment range to attempt to see more of a variation for field testing.
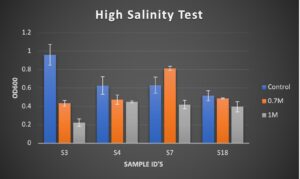
Figure 3. Results of OD600 measurements of bacterial isolates USDA 110, S18, and S16 after 3 days of growth. Treatments used were 0.0017M NaCl (Control), 0.7 M NaCl, and 0.5M NaCl.
Due to having a lower level of salt stress in the 0.7M NaCl range, the samples isolated from these glycerol stocks were sent for further study in the field trial experiment. Additionally, S3 isolate was added to the high salinity trial test after becoming contaminated during its medium salinity test. The S3 results for this test show salt stress at approximately 1M NaCl, but not 0.7M NaCl. Therefore, it was sent out to be used for field trial measurement in addition to the other listed samples. These results demonstrate that we see a broad range of response to abiotic stress and that we can isolate rhizobia with abiotic stress tolerance. The second phase was to take potential candidates to the greenhouse to impose stress in concert with a growing pea plant.
Greenhouse Trials
Isolates from the high salinity and N-plate fixing plates were grown in VR broth media and shaken (245rpm, 28 degrees Celsius) and diluted until they reached OD600=0.08. Treatments were subdivided thusly in equal concentrations, along with high nitrogen and low nitrogen solution control treatments.
Table 1. Treatment scheme for mass inoculations. Salinity Treatment = Week 1-2 ddH20. Week 3, 0.03M. Week 4, 0.06MNaCl, and 0.09M NaCl). Drought Treatment = Week 1, 100% H20. Week2, 100%, Week 3, 75% H20, Week 4, 50% H20, Week 5 25% H20.
|
|
Treatments |
||
|
Inoculant |
Salinity |
Drought |
H20 |
|
Inoculant 3 |
6 Replicates |
6 Replicates |
6 Replicates |
|
Inoculant 2 |
6 Replicates |
6 Replicates |
6 Replicates |
|
Inoculant 1 |
6 Replicates |
6 Replicates |
6 Replicates |
|
High Nitrogen |
6 Replicates |
6 Replicates |
6 Replicates |
|
Low Nitrogen |
6 Replicates |
6 Replicates |
6 Replicates |
Figure 4. Nodules observed from Inoculant 3.
Unfortunately, nodulation was only observed for Inoculant 3 (I.3), and there were correlations observed between I.3 and the other inoculants and the High Nitrogen control.3 had a higher average dry weight, as well as increased shoot length. Since further sequencing data points to plant growth promoting bacteria being a large source of isolates, this may be the attributed phenotypic properties observed. During harvesting on Week 6, only plants from I.3 contained observable nodulation. Their phenotype was observed to be very dark red/black nodules. These studies will be repeated to ascertain the reason for not getting nodulation after culturing the bacteria.
Field Observations
Lastly, the end goal of this project is to produce rhizobia that can be used in the field by farmers. Unfortunately, Covid created a number of obstacles in establishing field plots in 2020. We were unable to work with producers to establish our drought plots. However, we were able to at least set out a single site with several cultured rhizobia. The goal was to simply examine both N fixation and yield from two separate field pea varieties when inoculated with our native rhizobia species as compared to commercial granular and liquid inoculants.
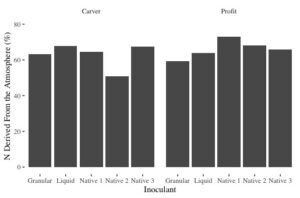
Figure 5. N derived from the atmosphere as a percent of total N in the plant across two field pea varieties (Carver and Profit)
As Figure 5 shows, the 3 native rhizobia inoculants performed similarly to the commercial granular and liquid inoculants. There was some variation between varieties, however, in general, all rhizobia fixed between 60-70% of N from the atmosphere. Likewise, total N fixed ranged from 60-80 lbs of N/acre, which was favorable to commercial inoculants. Hence, we these results are not conclusive, they do offer positive results. These tests will continue in 2021 with the addition of sites under saline and drought stress.
Figure 6. Total N (lbs/ac) fixed from the atmosphere across two field pea varieties (Carver and Profit)
2021 Results (lab results continued from 2020)
Objective 1: Rhizobia isolation.
Due to significant disruption from the Covid pandemic, much of our rhizobia screening was delayed until into 2020 and 2021. Due to the goal of isolating rhizobia that was resistant to high salinity environments, initially in this experiment’s conception, we chose the use of high salinity media broth as a “filtering” step before plating, choosing isolates based on morphology determination, and extracting dna for sequencing. However, this led to a number of false positives discovered after sequenced several isolates. Non-rhizobia isolates mostly composed of Pseudomonas and Baccilus strains. Because of this, a more stringent parallel plating system was used to first isolate candidate colonies before dna isolation. This system led to significant success in reducing the levels of false positives. And only after confirmation were the isolates then subjected to salinity stress test for further down-stream analysis.
Salinity-filtering Isolation
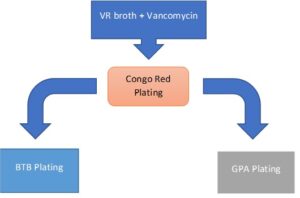
The following process was used for the initial phase of rhizobia isolation. Initially, samples were collected after incubation in 0.5M NaCl until post log phase, bacterial suspensions were plated onto vincent’s rich (VR) solid media supplemented with Congo Red (CR) (%1.2 Agar, 25 mg/L Congo Red). Positive isolates had favorable morphology (round, convex, glistening colonies) and color absorption from the CR. After 5 days of growth, samples with visible growth were replated on VR media. DNA was then extracted using DNeasy Ultraclean Microbial kit, as per instructions.
Parallel-plating Isolation
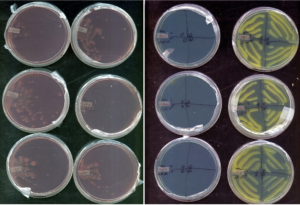
After false-positives were tested, an alternative method of isolation method was established, based on existing protocols (Working with Rhizobia Handbook, 2016). Samples were grown in VR liquid broth, with the addition of vancomycin antibiotic (concentration 1 mg/L), to limit contamination from gram-positive bacteria. After 3 days of growth, visible turgid samples were then streaked on CR plating for 3 days. Afterwards, colonies were chosen with favorable morphology, and further downstream parallel plating was added to differentiate rhizobia from possible non-rhizobia contaminants. Bromethyl bromothymol blue (BTB) (VR agar, 5 mL/L of 0.5% w/v in 0.2M KOH) plating and glucose-peptone agar (5 g glucose, 10 g peptone, 10 g agar) (GPA) plating was used. While colonies grow in media there are differentiations in phenotype on the plate based on pH-associated enzymatic reactions. While slow-growing rhizobia such as Bradyrhizobium, as well as contaminants such as Pseudomonas and Bacillus produce basic pH changes in the media during colony growth, fast-growing Rhizobium species produce an acidic reaction in the media. The addition of BTB dye to VR media produces a color change during pH shifts. Normally producing a green plate, BTB VR plates turn yellow when acidic and blue when basic. Additionally, GPA agar is used in medium where it is difficult for rhizobium to develop. BTB dye was added to the GPA agar to determine if contaminates are present. As can be seen, the test works as expected. The samples grew well on the CR plates as well as the BTB plates. In the latter, the colonies produced a yellow color that is described for an acidic reaction.
Figure 7. Plating scheme initially spreading colonies on Congo Red plates. Colonies without significant red contamination were streaked to BTB and GPA plates in quadrants searching for yellow coloring and no growth, respectively.
Figure 8. Scheme representation of parallel plating scheme. Bacteria isolates are cultured in VR broth supplemented with vancomycin gram-positive inhibitor. They are then spread on Congo Red plates, and round, convex, and glistening colonies (without significant red color absorption) are spread onto BTB and GPA plates concurrently. Samples that show yellow reactions in BTB plating, but no growth in GPA plating were made into a glycerol stock and used for sequencing.
Salinity Resistance of Suspected Rhizobia
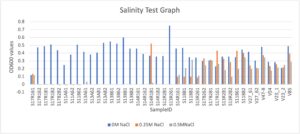
After successful plating, we conducted additional salt tolerance testing for rhizobia with potential drought stress conditions. A previous study had categorized tolerance scales for tested growth of microorganisms in saline conditions based on three groups: sensitive (0-50 mM NaCl), tolerant (100-500 mM NaCl) and extremely tolerant (600-1800 mM NaCl) (Pereira, 2008). Due to filtering potential helpful rhizobia, we determined testing scales of 0.00017M (control), 0.25M NaCl, and 0.5M NaCl, settling on an upper “tolerant” scale. While extremely tolerant upper limits of up to 1M was initially used for earlier testing, this had the potential to cause excessive filtering of helpful rhizobia species from the collected nodule glycerol stock cultures. Future tests exclusively used this scale for testing. Below is the results for salinity testing from the available rhizobia isolates following earlier screening and culling of the total candidate pool. As seen in Figure 9, the initial rhizobia isolates selected for greenhouse and field studies proved to be tolerant to high salinity, amongst several others. Overall, bacterial growth rate is considered to be in salt stress if there is 75% inhibition. Additional tests in the future might continue this work by testing further salt stress to test the limits of the bacteria’s osmotic stress, as well as an examination in the differences between genes governing abiotic osmotic stress.
Figure 9. Salinity test using bacteria isolates grown in three different scales of salinity,, 0M NaCl, 0.25M NaCl, and 0.5M NaCl. Samples were grown until post log phase, and bacterial suspensions of OD600 0.13, at a 1:100 ratio to VR. Samples were incubated for 5 days and bacterial pellets were centrifuged and resuspended in ddH20.
Rhizobia Identification
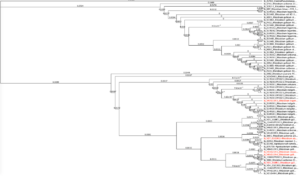
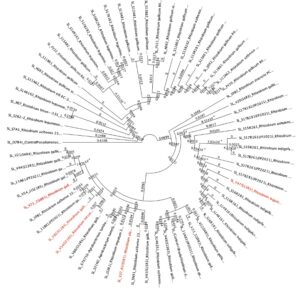
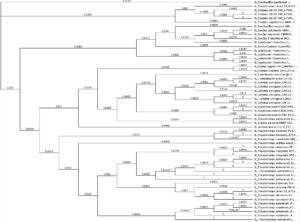
From the tests in Figure 9 (candidates on the right hand of the graph), we cultured rhizobia that were tested successfully for saline resistance in VR liquid media, and we extracted dna using DNeasy Ultraclean kits per specifications. We verified fidelity using Nanodrop 2000C. 16S primer pairs 27F and 519R were used for amplification. PCR protocol as follows (Degrees Celsius): Initial denaturation of 95 degrees for 4 minutes. 30x cycle of 95 degrees for 30 seconds, Annealing temp of 50 degrees for 45 seconds, and elongation phase of 72 degrees for 1 minute. And a final elongation phase of 72 degrees for 10 minutes. We verified pcr product using gel electrophoresis. After sequencing, samples were identified using BLAST (16S rDNA), and 5 candidate rhizobia strains were ultimately used for additional testing from collaborative greenhouse experiments. We uploaded sequence data to Geneious software, and we created phylogenetic trees from overall rhizobia collection results using Muscle (Tamura-Nei, Neighbor-Joining) for dna alignment and then tree building. The tree can be seen below. The highlighted red portions are the samples used in the field. In the Tree, a Pseudomonas strain, extracted from the same sample site and provided as a positive control for phenotype tests, as well as a well-characterized Glycine max-compatible Bradyrhizobium strain, was used as comparison points.
Figure 10. Phylogenetic trees, presented as rooted and circular trees.
Greenhouse Testing
Initial salinity testing, and sequencing of rhizobia isolates were used for field testing included V23.2, V2762, V47, V54, and V85, narrowed down from the original rhizobia isolates sequenced due to their high saline tolerance. Trials were ran to test for their effectiveness for growth in greenhouse settings. Similar inoculation strategy was used for all tested isolates in the greenhouse. Samples were grown in primary cultures of VR broth with ampicillin antibiotics (20ng/ml), and then post log phase were used to reinoculated secondary cultures until post-log phase, to ensure much reliable growth levels. Pea seeds were sterilized by being washed using 10% chlorax solution and 70% ethanol for 2 minutes each. They were rinsed with distilled water 10 times, and then placed in distilled water for 3 hours to imbibe.
Sterilized vermiculite/perlite mixture (3:1 ratio) were added in 1.5’’ by 8.25’’ cone-tainer cells. Pea seeds were added 1.5’’ below the fill line. Samples were watered with alternating ddH2O and ¼ PNS nutrient solution. After 1 week of growth, the samples were inoculated using cultured rhizobia. Two treatments were not inoculated with rhizobia, adding instead a nitrogenous Hoagland solution during the tray’s 1/4th PNS watering as a high nitrogen control. Another control had no rhizobia and no supplemental nitrogen source added. By comparing these controls, we can measure what value nitrogen sources supplemented by rhizobia. After 2 weeks of growth, the watering sources were supplemented with 0.03M NaCl. This value is increased an additional 0.03 until a final saline concentration of 0.09 NaCl was achieved. This value was chosen to stress the plant for abiotic conditions and test if the inoculated rhizobia was able to provide a mutualistic benefit by helping the plant survive under higher salinity conditions compared to the controls. These results showed a range of effectiveness for rhizobia isolated from the Midwest soils, both from their ability to impart biomass from saline resistance, as well as their capacity for measured nitrogen production.
Our goals in part focused on isolating these rhizobia for their ability to compete with market inoculants. There are many commercial inoculants that are offered in the market, that use a “one-size-fits-all” approach to develop high performing inoculants. In this trial, a commercial peat inoculant (CP) was added in comparison to the low and high nitrogen control. Salinity treatment was used for all samples. As can be seen from below, there was a range of effects observed for both saline and control treatment results. This was accomplished by, similar to the field, inoculating the pea seed in the same method as with inoculation in the field, simultaneously with rhizobia inoculation.
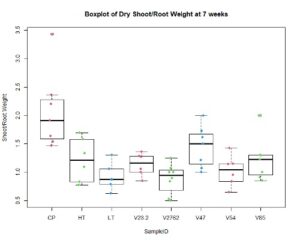
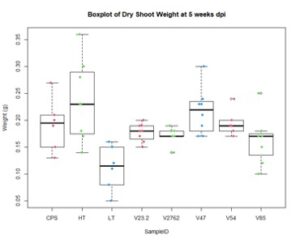
In the figures 11 and 12 below, we see the results for the non-saline treatments of the inoculated plants. And while V85 and V54 show a much higher distribution and mean (approximately by 3x) than the other V series, they are not statistically different at this level using Tukey test. Though when measuring shoot/root ratios, Peat control and V47 were statistically different than the other samples.
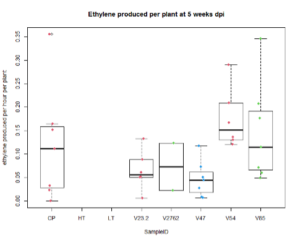
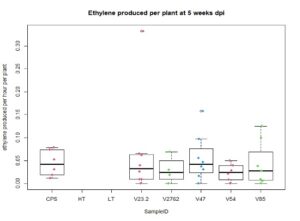
For the saline treatment (Figure 13 and 14), there was no significant difference observed between the ethylene production of the nodules measured. However, the Dry Shoot/Dry Root ratio showed that V47 provided the highest increase to shoot biomass distribution, with V2762 as expectedly the lowest alongside the Peat Control. This data showed that after saline conditions treatment, our rhizobia had a beneficial effect that was significantly greater than a negative control. Visual images also were important for this experiment, showing significant senescense and yellowing in the V23.2 and V2762 rhizobia pots. While this information gave conflicting information, it was useful to help narrow down focus for further field analysis. V2762 had difficulty producing nodules compared to the other inoculants, while previous work with V23.2 showed it to be less effective as well. While in this experiment it produced a significant amount of nodules, this was not notably producing significant amount of ethylene. The notable senescence present in those two sample pots were also a contributing factor for use in their long-term growth in the field.
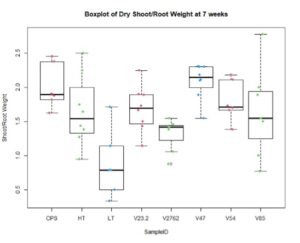
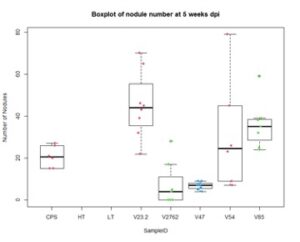
Further, nodule number under saline conditions varied significantly between rhizobia species, but these values do not necessarily correlate with biomass production. As Figures 15 and 16 show, V23.2 and V85 had relatively large numbers of nodules but middling shoot biomass, whereas V47 had very low nodule numbers but had the highest average biomass production.
Additional Rhizobia Isolation and Greenhouse Testing.
While screening for rhizobia samples initially for saline resistance, we potentially screened against possible rhizobia samples that might offer distinctive and beneficial phenotypes/mutualistic effects. There is an ever-growing body of research in not just single inoculant discoveries, but the examination of dual-inoculant studies that have shown increased growth benefits to legumes. By expanding our saline filtering step, we sought to explore the additional rhizobia strains that may have been initially filtered out, but could still have future capacity to provide mutualistic benefits in either single or dual-inoculant future experimental designs.
From our initial batch of testing, there were several strains listed with potential for very high nitrogenase enzyme production. Initial testing of these samples did not use salinity control, only as a biomass/ARA experiment. Downstream testing with these samples in mutualistic environments would be necessary to further match the overall goals of this project. If these samples are able to reproduce similar results under stress conditions, then there is potential for them to be used in further inoculant tests for field testing. Continuation of rhizobia study from isolated nodule samples is of potential significance for agricultural research.
Intrinsic Antibiotic Resistance
Testing for intrinsic antibiotic resistance (IAR) is necessary for culturing and testing bacterial isolates. Initially, ampicillin testing using liquid broth culture growth was conducted due to Rhizobium species having already widely established ampicillin resistance. However, we widened our tests for antibiotic resistance to gain more information on resistance plasmids, as well as to have multiple antibiotic options in case of potential ampicillin resistance. We used testing kits composed of antibiotic discs infused with ampicillin (10mcmg), chloramphenicol (30mcmg), streptomycin (10mcmg), and tetracycline (30mcmg). After culturing bacteria isolate in VR broth and resuspending the centrifuged bacteria pellet, sample turbidity was reduced to 0.13 OD600, and then 100ul of suspension was added to a VR solid plate. The suspension was spread using sterile glass beads. We separated the plates into quadrants, and antibiotic discs were aseptically affixed to the plate and stored in a 28 degrees Celsius incubator for 5 days. Afterwards, the halo-zone surrounding antibiotic discs, if any, were recorded (Figure 17,18). This provided a measure of the rhizobia’s susceptibility to tested antibiotics. The results of IAR for measured isolates that were used for downstream analysis is presented below. Notably, ampicillin remained the most consistent antibiotic for IAR. Samples were also widely resistant to chloramphenicol, but this resistance was inconsistent for several of the samples. Expectedly, all rhizobia samples had the least resistance to tetracycline, followed consistently by streptomycin.
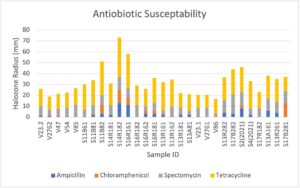
Phenotype Measurements
While salinity measurements provided a useful baseline measurement for stress resistance, there are many other sample tests that have been developed that are useful in establishing plant growth promoting effects.
IAA Production
Phytohormone secretion is one of the major ways in which soil bacteria are studied for their mutualistic benefits. Indole-3-Acetic Acid (IAA) is a very important phytohormone, a secondary metabolite of L-tryptophan that acts as a regulator of many biological processes for plant development while acting on organogenesis, trophic responses, and cellular responses such as cell expansion, division, differentiation, and gene regulation (Lebrazi et al., 2020). The production of IAA produced by colonizing bacteria in the plant rhizosphere proposed to act in conjunction with endogenous IAA in plant to stimulate cell proliferation, elongation, and enhancement of host’s uptake of minerals and nutrients from the soil (Shokri, 2010). The levels of IAA produced by a bacterium in the rhizosphere determines its effect on the host plant, with high levels inducing developmental abnormalities and stimulating formation of lateral and adventitious roots, while low levels promoting root elongation (Bal et al., 2013).
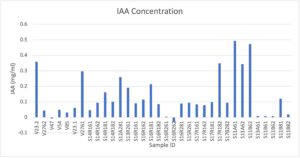
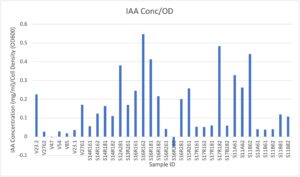
We sought to examine our rhizobia isolates for IAA production using previously established protocols. We cultured rhizobia isolates, resuspended bacterial pellet in VR broth, and reduced to 0.13 OD600. Samples were used to inoculate VR broth supplemented by 1 g/L of L-tryptophan. After 5 days of growth, these samples were removed, and centrifuged at 10,000g for 1 minute. 1ml of the supernatant was used to combine with Salkowski’s reagent (50ml 35% perchloric acid + 1ml 0.5M FeCl3.) at a 1:2 ratio. We set these samples in the dark and incubated them for 1 hour, as the chelating iron-based Salkowski reagent produced darker pink/purple colors with greater presence of IAA. Alongside, we used IAA standards combined with Salkowski reagent to estimate IAA concentrations. Afterwards, the samples were added to a flat-bottomed clear pcr plate, and the absorbances were read using a spectrophotometer set to 540nm.
The overall results for IAA experiments are presented below. Three graphs were presented here to demonstrate different measurements. The first graph shows results from calculations using the standard curve to derive concentration of IAA from the fluorescence values. The second graph shows a normalization of the concentration data to adjust for “cell number” of bacteria present in the final sample before IAA collection. The latter may serve to provide an idea of “efficiency” of the size of a culture Overall, these samples reveal lower levels of IAA produced by our field trial samples, especially V47 and V85. However, the V23.2 sample produced higher levels of IAA, and there was a wide range of IAA concentrations provided by our measured samples. Since IAA concentrations may be produced at different levels in under a variety of conditions, this is an exploratory snapshot that can allow us to dive further into other rhizobia of interest, as well as to compare how the experimental snapshot of this phenotype compares between sequencing data.
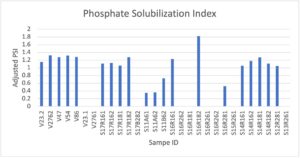
Phosphate Solubilization
Organic phosphates are widely present in soils but unusable by agricultural plants. While synthetic fertilizers are the typical alternative to soil organic phosphorus, this is, similar to nitrogenous fertilizers, a costly and polluting strategy in which only a fraction of used phosphate is absorbed into the soil. Many bacteria, however, do have the ability to convert organic phosphates in soil to a usable form by the legume. This is a PGPR that is favored by microbiology industries. Therefore, we used this as an additional test for our isolates to further score their effectiveness and potential future benefits as an inoculant product.
Using Pikovskaya’s agar we made a solid plate media contained with organic phosphates. We cultured isolates in VR, centrifuged at 10,000g for 1 minute, and resuspended pellet in sterile ddH2O. We reduced to 0.6 OD600, and spot inoculated solid media plates of Pikovskaya’s media. After 7 days of growth, we measured halo-zones surrounding colonies. The figure below shows the overall results of the phosphate solubilization tests conducted over our rhizobia isolates. The tested strains showed very similar levels of solubilization throughout the trials, though there were several samples that showed no solubilization under these conditions. Notably S16R182 showed the highest index levels.
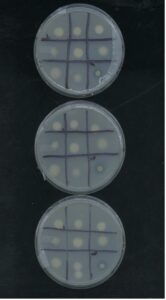
Isolation of non-rhizobia species
This project led to the identification of several non-rhizobia species from the crushed nodule glycerol stocks. There was a large number identified due to the initial false positives produced from the early isolation steps of the experiment (Figure 23). There was no significant phenotype testing from these bacteria due to falling outside the purview of our goals. However, notably Pseudomonas sample S7R4 provided a useful tool as a positive control for our several phenotype tests, including IAA, and phosphate solubilization testing. It was notable for outperforming a well-characterized phosphate positive control strain Pseudomonas aeruginosa strain ATTC 2853 in phosphate solubilization tests.
The use of non-rhizobia species, including Bacillus and Pseudomonas strains, as co-inoculants has shown to have beneficial cumulative effects. Non-rhizobial plant growth promoting bacteria has been shown to improve nodulation and grain yield of the legumes upon co-inoculation with crop specific rhizobia (Kohr, 2017). Therefore, these strains will be kept in storage and might provide further use for other researchers investigating co-inoculation effects. Below is a phylogenetic tree constructed from the non-rhizobia extracted isolates from the sample sites.
Field Studies - 2021
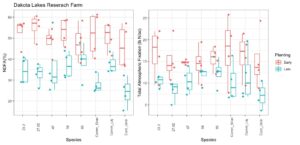
In 2021 we experienced drought conditions at both test sites. In western South Dakota (Sturgis) the earlier planting actually experienced more extreme drought conditions over the later planting date due to some late season rains. As figure 24 shows the late planting in general had higher nitrogen fixed from the atmosphere (NDFA) (as a percent of total N in the plant) and total nitrogen fixation. Overall, NDFA and total nitrogen fixation were roughly half of a typical year. Two native species - V47 and V85 - did comparably well against the commercial liquid and granular inoculants. Although the commercial granular inoculant had the highest N fixation, it was not statistically different from the two highest performing native rhizobia species.
At our second research site in the middle of the state, the drought was more severe and impacted the second planting more so than the first planting date. Figure 25 shows both the NDFA and total N fixation measured at this site. From a methodological standpoint, the uninoculated control (Cont_UnIn) was lower that than all other treatments at both sites, which suggests that our methods are free from cross-contamination. There is still native rhizobia in the soil at each research site, so some level of nitrogen fixation is expected. The results were not entirely consistent the with the other research site. However, V85 still performed quite well against the commercial controls. Lastly, Figure 26 depicts seed yield at both research sites. Again, overall yield was roughly 50% of long-term averages. There was not a consistent trend between treatments. V85 had the highest yield under the more mild drought stress conditions but did not fare as well under severe drought, despite having higher N fixation.
Overall, the 2021 lab and greenhouse studies increased our confidence in developing competitive rhizobia using native species. We have now developed a catalog of over 90 different rhizobia species and we have two very promising candidates from this pool. In 2022, we will continue our field studies for another year and expand to one additional salt-stressed field. Further we are in the process of obtaining the genetic sequences of several of our candidate species so that we can conduct an analysis of the different genes involved in conferring drought/salt tolerance. This work will allow us to streamline the rhizobia screening process with the possibility of identifying genetic markers for stress tolerance.
2022 Results
Field Studies -
In 2022, we repeated field the same field studies as in 2021. Two sites were established in Pierre, South Dakota and Sturgis, South Dakota at two planting dates (1. On-time and 2. Late planting). Unfortunately, the Sturgis site received several significant hail events during the growing season. While data were collected for this site, we did not feel that they were reliable due to the potential for inhibited growth effects by the hail. Hence, the data presented here is for the Pierre site only.
Nitrogen Fixation
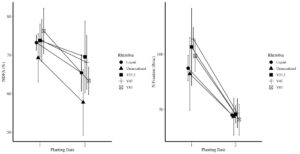
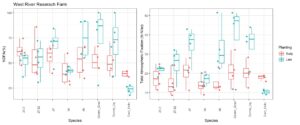
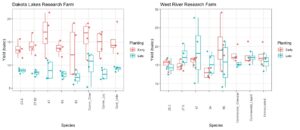
Contrary to 2021, precipitation in 2022 was much more conducive for plant growth. Both NDFA and total N fixation were correspondingly increased. On average, NDFA was 74% and total N fixation was 100 lb/ac for the on-time planting. For the late planting, average NDFA was reduced to 65% and total N fixation was 44 lbs/ac - a reduction of over 50%. Between treatments, the uninoculated control treatment had the lowest NDFA overall at 64% at the on-time planting and 58% at the late planting. As Figure 27 shows, the native rhizobia 23.2 had the highest NDFA at both planting times at 76% and 68%, respectively. Despite these differences, the inoculants were only statistically different (p<0.05) than the uninoculated control.
Similar to NDFA, total N fixation followed a similar trend although treatment differences were much greater at the on-time planting and were relatively similar at the late planting. In general, the native rhizobia fixed more nitrogen than either the uninoculated or liquid inoculant controls. V47 fixed the most nitrogen at 108 lbs/ac which compares quite favorably to the uninoculated control, which fixed 92 lbs/ac at the on-time planting (Figure 27).
Figure 27. The left-hand graph depicts the percent of total N in the plant derived from the Atmosphere (NDFA %) and the figure on the right depicts the total N fixed in lbs/ac by rhizobia treatment.
Yield
Similar to N fixation, yields were significantly decreased at the later planting date. On average, yields were 49 bu/ac at the on-time planting and just 17 bu/ac at the late planting. Within planting dates, yields were not statistically different between rhizobia inoculants although V23.2 trended lower, which suggests that this particular rhizobia may consume higher amounts of carbon than other rhizobia species (Figure 28). Moreover, this figure highlights the importance of inoculants for conserving soil N. As this figure shows, the uninoculated control yields were comparable to the other rhizobia inoculants. Assuming similar total nitrogen in the plant, the uninoculated control uses significantly more soil-derived N, which would leave less residual N for subsequent crops.
This project evaluated the use of native rhizobia as a means to fix nitrogen in field peas under abiotic stress such as drought. We developed a novel rhizobia screening protocol using increasing salinity culture to measure rhizobia growth. We hypothesized that rhizobia that could withstand high salinity levels would also be more drought tolerant as the physiological mechanisms to salt stress and drought stress are similar. To test this hypothesis, soils were gathered from unique ecosystems across the northern Great Plains (Montana, North Dakota and South Dakota). Fields peas were grown in each of these soils and the nodules that resulted naturally were used to extract the native rhizobia.
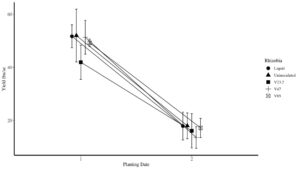
A total of 91 unique rhizobia species were cultured and initially characterized. Through salinity screening, we culled the list of potential candidates to 40 species, which were further subjected to lab tests including confirmation of rhizobia classification, further salinity tolerance, nitrogen fixation potential, nodulation potential and antibiotic screening (among others). From this pool, we identified 7 candidates that were advanced to field trials over two growing seasons. The experimental design including two testing locations with two separate planting dates. The idea for two planting dates was to include an 'on-time' planting and a 'late' planting date with the latter designed to move the critical flowering time (N-fixation maximum) into the later summer and the likelihood of higher temperatures and lower precipitation, hence placing greater stress on the plant-rhizobia complex. These two dates can then be compared to eachother with respect to total nitrogen fixation and yield between the native rhizobia species, commercial inoculant (positive control) and uninoculated field peas (negative control).
Across the two growing seasons, we experienced diverting weather conditions with severe drought persisting throughout the 2021 growing season at both sites and more favorable growing conditions in 2022. Unfortunately in 2022, one of our sites was lost due to hail, which left three site-years from which to evaluate native rhizobia performance. Overall, we were able to successfully demonstrate that native rhizobia screened using our salinity protocol can compete with 'elite' commercial rhizobia strands. However, while we saw some positive trends, the data remain inconclusive as to native rhizobia outcompeting commercial inoculants under stress conditions. For example, the percentage of nitrogen derived from the atmosphere (NDFA) in the plant averaged roughly 60% in all inoculated treatments and 53% in the uninoculated control at the on-time planting and approximately 57% in all inoculated treatments and 36% in the uninoculated control at the late planting.
Despite high variability, we did observe larger separation in total nitrogen fixation between rhizobia species, which is the product of the NDFA and total biomass. On average total N fixation was 43 lbs/ac at the on-time planting and 26 lbs/ac at the late planting. However, the range was a high of 108 lbs/ac to a low of 11 lbs/ac. The highest N fixing rhizobia was a native species cultivated for this project which averaged 14 lbs/ac more than the inoculated treatment and 17 lbs/ac more than the uninoculated treatment at the on-time planting. For the late planting, N fixation rates were similar with all inoculants fixing roughly 30% more than the uninoculated control.
With respect to yields, there was lower correlation between N-fixation and final seed yield. This suggests that there was adequate soil N to produce agronomically optimum yields. Indeed, the uninoculated control showed a seven bushel increase over the commercial liquid control at the on-time plantings. To be clear, the nitrogen derived from the atmosphere in the uninoculated control was generally well below the inoculated treatments. This implies that this treatment consumed far more soil-derived N than the inoculated treatments. Moreover, it may be expected that when N is limiting in the soil, yields would be significantly decreased for this treatment.
One exciting result from the yield analysis is that our strongest performing native rhizobia had yields as high as the uninoculated control and hence, also 7 bushels higher than the commercial inoculant. This is a major takeaway from this project because on average this rhizobia enables the plant to acquire over 60% of it's nitrogen from the atmosphere, which dramatically decreases the need for external N inputs. This rhizobia also fixed a high of 108 lbs/ac N at our Pierre location in 2022. Overall, this research demonstrates the great potential in using native rhizobia to create more resilient, climate-smart agricultural system.
Education
Our educational approach was based largely on in-field demonstrations and webinars. Covid precautions were in place for much of this project, which limited our ability for in-person events in the early phases of this research. As we were allowed, we brought producers and stakeholders into the field to see how these rhizobia work. Our education and outreach will continue and perhaps increase as we finalize our results and produce final conclusions beyond the official end date of this research.
Project Activities
Educational & Outreach Activities
Participation Summary:
Field days and demonstrations were described in the previous section.
This project also produced 1 PhD student and 1 Postdoc opportunity.
One peer-reviewed article:
Liping Gu, Jesus Raul Loya, Senthil Subramanian, Christopher Graham, Ruanbao Zhou,
Acetylene reduction assay for nitrogenase activity in root nodules under salinity stress,
Methods in Enzymology,
Academic Press,
2022,
,
ISSN 0076-6879,
https://doi.org/10.1016/bs.mie.2022.08.044.
(https://www.sciencedirect.com/science/article/pii/S007668792200369X)
Abstract: Nitrogenase, an enzyme present in a select group of prokaryotes reduces inert N2 into NH3 that can be utilized through biological pathways. This process, termed biological nitrogen fixation, plays a crucial role in the biogeochemical N cycle. The ability of nitrogenase to reduce acetylene to ethylene has been exploited to develop a reliable and accessible biochemical assay to measure this enzyme's activity. Biological nitrogen fixation by rhizobia bacteria that occupy root nodules of legume crops is a major source of sustainable nitrogen nutrition in agriculture. Environmental stresses exacerbated by climate change necessitate the need to evaluate nitrogen fixation in root nodules under various stress conditions. Here, we provide a detailed step-by-step protocol for nitrogenase activity measurements using acetylene reduction assay in field pea plants under saline stress. The protocol can be easily adapted for use with other biological systems.
Learning Outcomes
- The imperative of increasing crop rotations to include legumes/pulse crops
- Basic agronomic principles of pulse production
- The principles of nitrogen fixation
- Keys to successful plant-microbe symbiosis
- Production of rhizobia inoculant and efficacy of different inoculant formulations
- The value that native rhizobia can play in a more sustainable pulse production system under extreme abiotic conditions
- The potential for wild spaces to serve as genetic reservoirs for beneficial agricultural applications
- DIY methods for microbial inoculant production in the home lab
Project Outcomes
Inclusion of field peas in typical crop rotation
Use of granular inoculants and/or proper use of liquid inoculants
Diagnostic testing of effective plant inoculation/nodulation
One of the biggest and most rewarding successes from this project was through a workshop/presentation with the Northern Plains Sustainable Agriculture Society. This is a relatively tight-knit organization that caters largely to organic producers. Field peas are a principle crop in many of these farmers' rotations. The audience was very receptive to the research and excited about the opportunities that this work may bring in the future for new and improved rhizobia inoculants. Following the presentation and Q&A, we decided to make this workshop an annual event where we continue to build on previous work, discuss farmer success stories from the previous growing season and develop new DIY methods for producers to make their own inoculants.
There is significant opportunity to build upon this work, which we are hoping to do in the coming years.
- Explore specific genetic components that allow for native rhizobia to thrive under certain conditions
- We are currently in the process of sequencing the genome of the rhizobia we focused most of our research on including those we tested in the field. Unfortunately, these data were not back in time to include the analysis in this report.
- Develop methods for high-throughput screening of native/novel rhizobia
- One of the primary bottlenecks of this research was the ability to screen a large number of rhizobia species for stress tolerance. There was an even greater bottleneck in testing these rhizobia in the field. The fieldwork for this projects took an enormous amount of labor, time and space. It is extremely difficult to plant each rhizobia species in a statistically defensible manner while maintaining sterility (ie. preventing cross-contamination). Future research methods need to be developed to overcome some of the obstacles.
- Investigate the ability to co-inoculate either with multiple rhizobia species or with other beneficial microbes
- Two possibilities exists here:
- Can we combine different species with diverging or unique characteristics (perhaps one species very efficiently fixes N while another can survive extreme heat) to create a more resilient inoculant?
- When we sequence the full nodule genome, we generally find many other microbes along with the rhizobia. It is unclear why they are in this environment. Perhaps certain species are providing benefits to the plant and/or rhizobia that could be exploited.
- Two possibilities exists here:
- Explore opportunities to scale these processes to form a more distributed inoculant production capacity that could make inoculants tailored to an individual farmer's field
- Moving beyond the one size fits all model to an industry build on custom inoculants
- Determine the survivability of native rhizobia compared to commercial products
- We do not have a good sense for how well the native rhizobia compete for nodule occupancy and if they can maintain high populations without continued inoculation vs. a commercial product.