Final report for SW15-015
Project Information
The goal of this project was to validate previously-identified genetic markers shown to be associated with cattle use of rugged terrain and areas far from water. We hoped to verify and enhance prototype DNA test(s) that could be used to identify cows and bulls with superior and inferior grazing distribution genotypes. Potentially, these tests can be used to identify bulls that will sire daughters that use more rugged topography and travel farther water.
Cattle tracking data and DNA samples were collected at eight ranches in New Mexico, Colorado, Nevada, Arizona and Wyoming and combined with data collected from our previous Western SARE study (SW09-54) for a total of 330 cows. BovineSNP50 genotyping data were used to explain tracking data using the BOLT software package with BayesC methodology. Quantitative trait loci detection was based upon posterior inclusion probability (PIP) and the percentage of total genetic variance explained by individual genomic windows. Analyses revealed 32 quantitative trait loci (QTL) that were previously associated with health and production traits in dairy and beef cattle as well as 29 putative candidate genes that function in oxygen homeostasis, environmental adaptation, and feed efficiency and growth. The QTL detected in this study support the polygenic nature of complex traits regulating beef cow terrain-use. Detection of genotype to phenotype associations in the current study was likely limited by the moderate sample size and lack of uniformity in the data. Therefore, a large independent population of beef cows, composed of one breed, grazing on the same pastures is needed to refine terrain-use measurements and further elucidate the role of genetics in terrain use in beef cattle.
Starting in December 2018, results and conclusions from this study as well as other grazing management tools were presented to over 620 ranchers and land managers at a total of 13 meetings located in New Mexico, Arizona, Nevada and California. A scientific symposium focusing on this project was presented the Society for Range Management 2019 Annual Meeting in Minneapolis, MN that was attended by over 60 scientists and land managers.
A. Validate previously identified genetic markers discovered from previous Western SARE funded research project (SW09-054) shown to be associated with cattle grazing distribution.
A1. Track at least 200 cows with GPS collars for at least 2 months at 7 or more mountainous ranches in New Mexico, Arizona, Colorado, and Nevada.
A2. Collect DNA samples from tracked cows.
A3. Analyze DNA from tracked cows using High Density single nucleotide polymorphism (SNP) chip (BovineSNPHD; ~770,000 genotypes) and determine if previously identified genetic markers are similarly associated with grazing distribution traits as found in the previous Western SARE study SW09-054.
B.Verify and enhance prototype DNA test(s) for identifying superior and inferior genotypes for grazing distribution.
B1. Analyze genetic markers used in a small prototype (180 SNP) panel with cows tracked in this project and compare associations to previously studied cattle herds.
B2. Add or replace genetic markers in the prototype SNP panel so that the most predictive SNP are used in final DNA test.
C. Develop procedures and end user tools for estimating molecular breeding values (MBV) for grazing distribution.
C1. Develop statistical procedures for estimating MBV for grazing distribution from cattle DNA samples.
C2. Automate download of genotypes from DNA service providing company into CSU Center for Genetic Evaluation of Livestock (CGEL) software for estimating MBV.
C3. Develop web-based software so producers can easily obtain the MBV for grazing distribution for each animal.
D. Provide training to ranchers and seedstock producers specifically designed to increase their knowledge and ability to utilize MBV to select cattle for improved grazing distribution.
D1. Show ranchers how to collect DNA samples from cattle and how to submit the samples for analysis.
D2. Teach ranchers how to obtain MBV from web-based systems for their cattle and utilize this information to select for improved grazing distribution.
D3. Show how ranchers can incorporate grazing distribution goals and objectives into their ranch-specific breeding program.
E. Estimate the potential for improvement of grazing distribution through MBV and genetic selection.
E1. Estimate the potential for improvement in grazing distribution through genetic selection under varying scenarios.
E2. Deliver this information to ranchers and land managers via multiple kinds of extension programs so they will know the potential for improving on-the-ground grazing use.
Cooperators
- - Producer
- (Researcher)
- - Producer
- - Producer
- (Educator and Researcher)
- (Researcher)
- - Producer
- (Researcher)
- (Researcher)
- - Producer
- - Producer
- (Researcher)
- - Producer
- (Educator and Researcher)
- - Producer
- - Producer
Research
Our previous SARE study (SW09-054) showed that grazing distribution could be inherited and demonstrated the potential for practically applying selection to manipulate cattle grazing patterns. However, these results were based on 160 cows and additional research was needed before those exciting results could be used by ranchers. We proposed to validate previously-identified genetic markers shown to be associated with cattle use of rugged terrain and areas far from water. We hoped to verify and enhance prototype DNA test(s) that could be used to identify cows and bulls with superior and inferior grazing distribution genotypes. Potentially, these tests could be used to identify bulls that will sire daughters that use more rugged topography and travel farther water. These DNA tests could also be used to select (cull) cows with superior (inferior) genotypes. Such selections would not require expensive GPS tracking. Only a blood sample and DNA test would be needed at a cost of potentially less than $30 per animal. Data from DNA tests are complex so we planned develop molecular breeding values (MBV) for producers to use in their cattle breeding selection programs. The MBV would be similar to routinely calculated Expected Progeny Differences (EPD’s) that ranchers regularly use to select bulls and replacement heifers.
. Global Positioning System (GPS) tracking data and DNA samples were collected at 12 ranches to verify results of our previous research project (SARE SW09-054). The ranches are 1) Heartstone Angus (Dick and Erin Evans) near Silver City, NM; 2) Wilbanks Ranches (Aaron Wilbanks) near Mayhill, NM; 3) Ensz Ranch (Wesley Ensz) near Center, CO; 4) Silver Spur Ranches (Nick Wamsley) near Saratoga, WY; 5) Humboldt Ranch (Jessie Braatz) near Paradise Valley, NV; 6) Gund Ranch (Jon Wilker) near Austin, NV; 7) the OR O Ranch near Prescott, AZ, 8) NMSU College Ranch (aka Chihuahuan Desert Rangeland Research Center) near Las Cruces, NM; 9) Fort Union Ranch (James Stuart) near Watrous, NM, 10) Egan Ranch (Joe Eagan) near Susanville, CA, 11) Roberti Ranch (Rick Roberit) near Vinton, CA, and 12) John E. Rouse Beef Improvement Center (Rouse Ranch, Colorado State University) near Encampment, WY. We tracked 15 to 35 randomly-selected mature Angus or Angus cross cows at each ranch. Cows were tracked in extensive pastures with rugged terrain using GPS collars that recordied positions at 10-minute intervals for 2 to 5 months at each ranch. Blood samples were collected from all tracked cows. DNA was stored until genotyping analyses using commercially available free-to-air (FTA) cards GPS tracking data, digital elevation models, and geographical information software were calculate terrain use metrics based on our previous study. Indices were used to combine individual cow use of slope, elevation and distance from water so that cows can be ranked and compared at each ranch (contemporary group analyses).
All tracked cows from this study were genotyped using the Illumina Bovine SNPHD array, which evaluates approximately 770,000 genetic markers (i.e., single nucleotide polymorphisms; SNP) Samples were genotyped using an allele discrimination platform by MALDI-TOF mass spectrometry (Sequenom MassARRAY(R)) (Neogen/GeneSeek Inc., Lincoln, NE) to perform association studies. We were able to track and genotype more cows than we originally planned. However, we did not have time to include the results from the last 80 cows in this final report. We continued to track cows and obtain DNA samples throughout the entire study because a large sample size is critical in genetic studies. The data from these additional 80 cows will be used in future planned analyses.
Data from 37 cows from the Rouse Ranch that were not included in this study or the previous Western SARE study (SW09-052 or SW15-015) were genotyped with the BovineSNP50 Beadchip (53,714 SNP). The high-density SNP data were truncated to match the BovineSNP50 data to generate cohesive genotype data for analyses (n = 330). Genotype quality control was completed using PLINK 1.9 software and standard filters. Genotype data were pruned for linkage disequilibrium (LD). Relatedness of individuals and population structure were evaluated using identity by descent and principle component analysis (PCA). First and second degree relatives (pi‐hat > 0.2) were retained in this study (n = 154) to maintain the sample size. A PCA plot, generated using RStudio (version 3.3.2), did not suggest population stratification (i.e., subpopulations with systematic differences in allele frequencies due to ancestry) as individuals did not form distinct clusters. Therefore, after quality control, 321 animals and 42,603 SNP were available for the vertical climb analysis and 323 animals and 42,699 SNP were available for all other analyses. Genotypes were recoded from AB format to numerical values (AA [-1], AB [0], and BB [1]) and missing genotypes were filled with the median for that locus.
A genome wide association study was conducted for each phenotype using the BOLT software package (Release 1.2.7) and BayesC methodology. Given the difficulty of measuring terrain-use across western U.S. rangeland ranches, numerous fixed effects were examined during model development including: ranch, breed, terrain type (mountainous or rolling), season (spring, summer, fall, winter), physiological status (lactating or dry), collar type (Lotek or igotU), and the GPS tracking start date. Backward selection with alpha 0.05 was used to identify significant predictors for each model. All of the predictors, except collar type, were linearly dependent with GPS tracking start date and therefore, these variables were removed from the models. Collar type was not significant (P > 0.05), and thus the most appropriate model included GPS start date as a fixed effect. Fitting start date in the model was analogous to fitting a designated contemporary group to account for environmental differences among cows on ranches and pastures.
Allele substitution effects, variance of the allele substitution effects, and the number of times the marker was included in the model (count) were derived using a single-site Gibbs sampler with 150,000 iterations. Posterior inclusion probability (PIP) or the proportion of iterations in the Markov chain Monte Carlo (MCMC) chain that included a particular SNP was calculated for each marker and the five SNP with the highest PIP were identified for each phenotype. These SNP were termed the candidate SNP throughout the study. Cattle QTL database (Cattle QTLdb; https://www.animalgenome.org/cgi-bin/QTLdb/BT/index) was used to determine if the QTL had been previously associated with any beef cattle traits. The Ensembl genome database (Release 94) was used to identify genes within one megabase of the candidate SNP and the annotated gene located nearest to SNP was deemed the putative candidate gene. Gene ontology was examined using AgBase (version 2.00; http://agbase.arizona.edu/index.html).
Bayesian multiple-SNP regression may fail to reveal strong associations between individual markers and the trait of interest (i.e., low PIP or small percentage of genetic variance explained) due to linkage disequilibrium; therefore, associations are often identified using genomic windows. For this study, non-overlapping one megabase genomic windows were derived using the annotation for bovine assembly UMD3.1.1. The MCMC sampling chain values were used to calculate the proportion of genetic variance explained by the markers in the genomic windows and the five genomic windows explaining the largest portion of genetic variance were deemed regions of interest. The SNP with the highest PIP within the region of interest was considered the lead SNP and Ensembl genome database (Release 94) was used to identify the annotated gene located nearest to the lead SNP. Beef cattle traits previously associated with the QTL were identified using Cattle QTLdb (https://www.animalgenome.org/cgi-bin/QTLdb/BT/index) and AgBase (version 2.00; http://agbase.arizona.edu/index.html) was used to assess the gene ontology.

Results from Genome Wide Association Analyses. Five SNP were associated with each of the six terrain-use traits: slope, elevation, vertical climb, distance from water, rough index, and rolling index. Several SNP were associated with more than one trait and therefore, 26 unique SNP effects were detected. Single nucleotide polymorphisms associated with terrain-use traits were observed on chromosomes 1, 4, 5, 8, 10, 11, 12, 13, 16, 17, 18, 21, 22, 23, 24, 27, and 29, though the largest number of variants was found on chromosome 10 (n = 5). The posterior inclusion probabilities for these 26 SNP ranged from 0.02 to 0.18 (Figure 1). Marker rs41600226 on chromosome 23, associated with elevation, had the highest PIP across all association analyses. The mean PIP for all SNP considered was 0.006 for elevation and vertical climb whereas the mean PIP for slope, distance travelled from water, rolling index and rough index was 0.005.
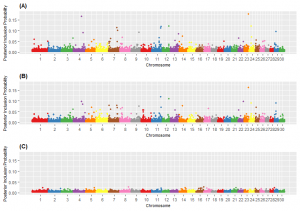
Of the 26 SNP that were associated with terrain-use traits, 10 were intronic variants and 16 were intergenic. Further examination of the candidate SNP using Cattle QTL database revealed that 24 QTL had been previously identified and associated with production and health traits in dairy and beef cattle including: birth weight, carcass weight, residual feed intake, milk yield, and bovine tuberculosis susceptibility. Putative candidate genes were related to diverse biological functions including: gene expression, intracellular transport, and glucose metabolism.
Genome Wide Associations Analyses Discussion. The genome-wide association study revealed QTL that may be important for terrain-use in extensive rangeland pastures. The posterior inclusion probabilities for the SNP in the six GWAS were considered low; however, Manhattan plots for five of the six traits had notable peaks and a review of the literature revealed no standard threshold for determining significance based on PIP. Furthermore, previous mammalian studies suggested that PJP varies greatly depending upon the trait of interest. In a previous study, cooperators in this study (Speidel et al. 2018 J. Anim. Sci. 96, 846-853) examined the genetic architecture of Heifer Pregnancy and Stayability in Red Angus cattle and set a PIP threshold for each trait. Speidel et al. (2018) reported lower PIP for Heifer Pregnancy (6%) than Stayability (100%). More specifically, the maximum PIP for heifer pregnancy was 6% whereas the max PIP for stayability was 100%. These authors hypothesized that the low inclusion of markers for heifer pregnancy was due to limited phenotypic data on genotyped individuals (n = 567) and the low heritability of the trait (h2 = 0.12). Perhaps, the PIP for the terrain-use traits presented in this study were influenced by similar factors. The moderate sample size of this study limited the statistical power for QTL detection and results suggested that terrain-use traits may be lowly heritable. Yet, this study was challenged by landscape diversity among ranches, breed of cattle, and our ability to adequately quantify the difficulty cattle incur when utilizing steep slopes, climbing and descending rugged topography and traveling long distances from water. Terrain use is a multi-dimensional trait that should integrate slope, vertical climb (or elevation) and distance traveled from water. Although the rough and rolling terrain indices combine these traits into one value, alternative approaches to integrate slope, vertical climb and distance traveled from water into a single trait deserves further study.
Twenty-four genomic windows, spanning 13 chromosomes, were associated with the six terrain-use traits and six of the windows were associated with multiple traits. Within the genomic windows, 25 lead SNP were identified. Parallel with the PIP, the genomic windows explained a low proportion of genetic variance for the terrain-use traits. Across the six traits, the maximum proportion of genetic variance explained by a single window was 0.0057%. Again, these estimates may have been influenced by the heritability and marker density (i.e., the number of markers captured in the genomic window). A greater proportion of genetic variance can be explained using dense genotyping arrays as there is greater linkage disequilibrium between causative mutations and the surrounding markers. Another important consideration regarding the proportion of genetic variance explained was the genetic architecture underlying quantitative traits. Complex quantitative traits that are influenced by many QTL with small effects (i.e., QTL explain a small percentage of total genetic variance on an individual basis. Genomic regions containing causative mutations may encompass few SNP with large effects whereas the remaining regions contained many markers with small effect.
A comparison of the candidate SNP to the genomic windows revealed 32 QTL and 29 putative candidate genes that may play a role in beef cow terrain-use. Thirty of the QTL were documented in Cattle QTLdb and associated with numerous beef cattle production traits. The 29 putative candidate genes identified in this analysis were not concordant with the five genes identified by our previous Western SARE study SW09-054 (Bailey et al. 2015, Rangeland Ecol. Manage. 68, 142-149). Confirmation of the previously identified genes may have been hindered by differences in SNP density (high-density genotypes vs. BovineSNP50 genotypes), statistical methodology (single-SNP regression vs. Bayesian multiple-SNP regression), and extreme topography variation among rangeland beef cattle operations as new data were added to these resources. Loci on chromosome 29 were detected in both studies and warrant fine mapping efforts to understand and identify the potential causal mutations.
Our previous study Western SARE 09-054 (Bailey et al. 2015) used BovineHD genotypes and single-SNP regression analysis to identify QTL associated with beef cattle terrain-use. Significant markers identified in this analysis were used to develop a custom-genotyping panel of 50 SNP genotypes for subsequent association analyses with phenotypes. The genotype data in this study were derived from BovineSNP50; therefore, the eight candidate SNP discovered by our previous study (SW09-054, Bailey et al. 2015) were not replicated in this study. In our study, Bayesian methodology was used instead of single-SNP regression because simultaneously fitting markers accounts for all SNP in linkage disequilibrium (LD) with the QTL which decreases the proportion of unexplained genetic variance. In addition, Bayesian multiple-regression results in fewer false positives because population structure was explained in the model. Differences in the genomic architecture (i.e., allele or genotypic frequencies) between study populations (ranches) most likely influenced QTL detection.
All of the ranches except Carter Ranch were in moderate (3,940 to 5,250 feet) to high-elevation regions (≥ 5250 feet) where a reduction in atmospheric pressure results in lower partial pressure of oxygen. This may help explain the association between the previously discussed candidate genes and the terrain-use traits. Cattle that are utilizing rugged terrain in high altitude regions may experience hypoxia as oxygen consumption increases in active muscle cells. In hypoxic conditions, mammals may undergo physiological changes to compensate for the lack of oxygen including angiogenesis, erythrocyte modification, heart remodeling, and accelerated gluconeogenesis. Furthermore, cattle maintained in high-elevation regions may develop high altitude disease as a result of hypoxia induced pulmonary arterial hypertension. The role of the hypoxia candidate gene and the relationship between pulmonary arterial pressure and terrain-use of cattle deserves further study.
As previously discussed, three of the putative candidate genes (INHBA, LRRC4B, and SEPT7) were associated with growth traits and feed efficiency in cattle. The functions of INHBA, LRRC4B, and SEPT7 are particularly interesting given the potential relationship between cow-size and terrain-use and locomotion and residual feed intake (RFI). Use of rugged terrain-use should require greater energy expenditure and be unfavorable for low RFI. Additional studies with an independent population are needed to further examination the relationship between mature cow weight/size, residual feed intake, and terrain-use traits.
The QTL detected in this study support the polygenic nature of complex traits regulating beef cow terrain-use traits. Detection of genotype to phenotype associations in the current study was likely limited by the moderate sample size and lack of uniformity in the data. Therefore, a large independent population of beef cows, composed of one breed, grazing on the same pastures is needed to refine terrain-use measurements and further elucidate the role of genetics in beef cattle terrain-use phenotypes.
Analysis of Previously Identified Candidate Genes. Our previous study (SARE SW09-54) suggested that the traits used in indices to quantify grazing distribution are moderately heritable; therefore, genomic selection could be used to improve grazing distribution. Five quantitative trait loci (QTL) and underlying candidate genes (ACN9, FAM48A, GRM5, MAML3, and RUSC2) were found to be associated with grazing distribution traits in cattle. We examined these candidate genes and identified additional SNP that can be incorporated into a previously developed 50-SNP panel used for genotype associations with grazing distribution phenotypes (Western SARE SW09-054). Sequencing of RNA (RNA-Seq) yielded 30 million reads (single-read) per sample from 6 tissues (aorta, LM muscle, lung, pulmonary artery, and right and left ventricle) collected as part of an altitude tolerance study of Angus cattle. These tissues were from 10 steers with outlying pulmonary arterial pressure observations and unique sires. Sequences were assembled to the annotated bovine reference genome (UMD3.1; release annotation 87) and analyzed using CLC Genomics Workbench (version 8.0). Variant detection was performed using two methods: (1) individual samples and (2) a pool of all samples. No variants were detected in GRM5, MAML3, and RUSC2; however, individual sample analysis identified 30 SNP within ACN9 and FAM48A, and pooled sample analysis identified 184 SNP within ACN9 and FAM48A. Twenty-one SNP were identified in both approaches. The Ensemble Variant Effect Predictor was used to determine the functional consequence of each SNP. Of the 21 SNP, 16 were intronic, four were exonic, and one was reported to be a downstream variant and a splice acceptor variant. The SNP discovered using RNASeq technology were compared to the exonic SNP in dbSNP within the 5 candidate genes. There were 1,663 exonic SNP in dbSNP in these genes. One synonymous SNP, located within ACN9 (rs382949979), was observed in both data from RNASeq and dbSNP. In summary, 21 SNP were discovered in two of the five candidate genes underlying QTL associated with grazing distribution that were identified in the previous study (SW09-054).

Terrain Use Indices. Livestock grazing distribution is a complex trait, which makes it difficult to evaluate in genome wide association studies. Slope and horizontal and vertical distance to water, alone and in combination, affect how cattle use mountainous rangelands, which makes terrain use a multidimensional trait. Assessment of terrain use by cattle is difficult because the metrics used to describe aspects of terrain use different scales of measurement. For example, slope is measured in percent or degrees, while distance from water is measured in meters. Ongoing studies are developing indices that can be used measure and evaluate terrain use of cattle n rugged rangeland. Tracking data from 15 to 18 cows at each of 4 ranches in New Mexico, Montana and Nevada were used in resource selection analyses. The ranches had different terrain types varying from rolling to very mountainous terrain. Slope, elevation, and distance to water at overlapping 100-m diameter randomly-selected plots were calculated at each ranch. Cattle use of each plot were used general linear model with a negative binomial distribution. Feature scaling was used to normalize these variables so that could be evaluated with relatively similar units. Except for the ranch with rolling terrain, cattle selected again steep slopes (linear relationship). Cattle selected against higher elevation in mountainous terrain, and the relationship was quadratic. Cattle selected again areas far from water at all ranches, but the relationship was quadratic rather than linear. These results suggest that feature scaling and resource selection analyses may be tools to help evaluate the multidimensional aspects of terrain use in cattle, but the relationships between terrain metrics will likely vary among ranches because of the unique terrain characteristics of each pasture.
Repeatability of Cattle Grazing Patterns. We used tracking collars to monitor cattle movements and evaluate grazing distribution phenotypes. However, few studies have evaluated the consistency of cattle movements over time in rugged rangeland pastures. We examined the repeatability of cattle movement patterns at five locations in New Mexico and Arizona: the Chihuahuan Desert Rangeland Research Center (CDRRC), Evans Ranch, Wilbanks Ranch, Hartley Ranch and Todd Ranch. These data were obtained from this study and our previous SARE study (SW09-054). Eight to 19 randomly-selected cows from herds of 40 to 200 cows were tracked with GPS collars at each ranch. Cows were tracked at either 10- or 15-minute intervals. Terrain use was summarized by week. A repeated measures analysis was conducted on each ranch using the weekly average of slope use, elevation use, and distance from water as the dependent variables. Intraclass correlations of weekly averages of the three terrain use metrics were used to assess temporal consistency of grazing distribution traits. Week was a fixed effect and cow was a random effect. Intraclass correlations of terrain use by individual cows varied among ranches. The Wilbanks Ranch had the strongest intraclass correlations for slope, elevation, and distance to water of 0.60, 0.50, and 0.77, respectively. Intraclass correlations for elevation at the Hartley and Todd Ranches were strong, 0.61 and 0.71, respectively, but correlations for slope and distance to water were weak to moderate (0.18 to 0.30). In contrast, intraclass correlation at the CDRRC and Evans Ranch were weak (0.00 to 0.08). Our results suggest that consistency of terrain use by cattle can vary by location; however, these relationships are positive and moderate to strong at most ranches. Factors such as cattle familiarity with pastures and the nature of the terrain features may explain part of this variability, however additional analyses examining how temporal changes in terrain use affect this phenotype are needed.
Terrain use and grazing distribution traits in cattle are difficult to evaluate because movement patterns are temporally variable due to ever changing climatic and forage conditions. Terrain-use can be monitored with GPS collars, but the length of tracking is constrained by battery life and on-site grazing and management plans. Cattle tracking studies have varied from days to months. We evaluated data from our previous SARE study (SW09-054) to determine if shorter, 30 to 60 day, sampling durations were as effective as a 3 month tracking periods for characterizing slope and elevation use, and vertical and horizontal distance traveled from water of individual cows. Fifteen Limousin cows from a herd of 250 cows were tracked at 15-minute intervals for 92 days during late winter and early spring in a 9,065 ha pasture that included both gentle and rugged terrain. Terrain-use during 30- and 60-day periods at the beginning and end of the tracking were compared to the full 92-day period. Slope, elevation, and vertical and horizontal distance from water from the full sample period were regressed on values from the shorter data subsets and correlations were calculated between sampling periods for each pair of terrain use metrics. The 60-day periods showed strong agreement with the full 92-day period with correlation coefficients varying from 0.90 to 0.97. Correlations between 30-day periods and the 92-day period varied from 0.30 for distance to water during the early period to 0.90 to 0.93 for all traits during the late 30-day period. These preliminary analyses suggest that 2-month tracking periods are equivalent to 3-month tracking periods for identifying differences in terrain-use among beef cows. These findings help facilitate collection of grazing distribution phenotypes from larger number of cattle for genetic improvement.
Only a few studies have examined the impact of cow size on grazing distribution. Tracking data from 74 mature cows collected in this project at the CDRRC (n = 14), Silver Spur Ranch (n = 28) and Fort Union Ranch (n = 32) were used to determine if there are relationships between cow body size and terrain use. After fitting the statistical model for ranch, residual correlations were used to examine the relationship between cow body condition score, linear measures of cow size, and terrain use metrics. No important residual correlations were detected between terrain use metrics and cow body measures (P > 0.05). These results suggest that terrain use of cattle is not related to cow size and body condition. Although genetic correlations need to be evaluated, the phenotypic correlations suggest that selection for terrain use should not affect cow size or body condition.

Genome-wide association analyses using Bayesian approaches and data from 330 beef cows managed on 14 ranches revealed 32 SNP and 29 putative candidate genes for terrain-use. Many of the QTL were previously associated with beef and dairy cattle health and performance traits. Four of the 29 putative genes lacked functional annotation; however, the remaining 25 genes were related to a variety of biological processes including hypoxia, feed efficiency and weight traits.
Research Outcomes
Education and Outreach
Participation Summary:
While the study was being conducted, we presented results from our previous Western SARE study (SW09-054) and a summary of this study at two international conferences: International Rangeland Congress in Saskatoon, Saskatchewan, Canada (July 2016) and the PA17 – The International Tri-Conference for Precision Agriculture in 2017 in Hamilton, New Zealand (October 2017). Results were also presented scientific conferences in the USA: Society for Range Management Annual Meeting in February 2016, January 2017 (St George, UT) and January 2018 (Reno, NV and the American Society of Animal Science in July 2017 (Baltimore, MD). An invited presentation focused on this project was given at the Beef Symposium at the Western Section of the American Society of Animal Science Meetings during June 2018 in Bend, OR. Two invited presentations summarizing this project were presented at the Beef Improvement Federation during June 2018 in Loveland, CO.
As the study came to an end (since December 2018), presentations focusing on this project were given to ranchers and land managers at 12 locations in Arizona (3), California (1), Nevada (6) and New Mexico (2). In addition we presented a day-long workshop for producers in Chico, CA based entirely on this project in February 2019. We also held a 1/2 day symposium summarizing the results of this project at the Society for Range Management Annual Meeting in Minneapolis, MN.
We published a University of Arizona Cooperative Extension fact sheet (az1760-2018) based on this project and our previous Western SARE project (SW09-054) in March 2018 entitled "Nature and Nurture's Influence on Cattle Distribution. We have just submitted a journal article to Livestock Science based on results from this study entitled "Genome-Wide Association Studies of Beef Cow Terrain-Use Traits Using Bayesian Multiple-SNP Regression." We plan to submit at least 2 more journal publications based on the results of this study.
Education and Outreach Outcomes
In the future, we plan to continue this research so that we can complete our goal of developing a reliable and cost effect breeding value (e.g. EPD) for beef cow terrain use. In the next study, we plan to track at least 400 mature cows of the same breed in the same mountainous pasture for at least 2 months. Ideally, the cows would be purebred and have pedigrees. In the past, tracking 400 plus cows in the same pasture would be cost prohibitive because the GPS collars were too expensive. At the beginning of the study, reliable GPS collars were $1800 each, but we developed a low cost collar that was $250. However, we are now working with a company that plans to have an ear tag with GPS that can track cow for about $40 each. With the lower cost of GPS tracking units, we can now track several hundred cows in the same pasture at the same time. We believe this will solve most of the analysis issues that we encountered in this study.
- Genetic selection
- Livestock grazing management
Livestock grazing management
Genetic selection