Project Overview
Annual Reports
Information Products
Commodities
- Additional Plants: ornamentals
Practices
- Education and Training: extension, workshop
- Pest Management: chemical control, cultural control, disease vectors, eradication, field monitoring/scouting, integrated pest management, physical control, prevention, sanitation
Abstract:
Little research has been done to diagnose the causal agents of various diseases that affect peony. Modern extension bulletins and growers’ guides rely on nearly 100 year-old research to diagnose disease on this high-value cut flower and landscape crop. In order to provide growers with more information, samples of peonies were collected from 12 states in the United States during the summer of 2016 with the purpose of identifying various pathogens causing disease. Fungal plant pathogens were isolated from diseased tissue and identified by morphology and PCR and sequencing of various informative genes. A total of 10 fungal genera were identified, including five genera never before reported on peonies in the United States: a Botryosphaeria species, multiple Colletotrichum species, Mycocentrospora acerina, a Phoma species, and Pilidium concavum. In addition to those new to the United States, a number of new state-pathogen-host combinations were identified in the survey. A list of the state-pathogen combinations found as a result of this survey, combined with data from previous years that helped justify funding for this grant are displayed in Table 1. Koch’s postulates were performed for most first reports to confirm pathogenicity. The information gained from this survey will be valuable for growers and disease diagnosticians to make better diagnoses and subsequent disease management decisions. Additional diagnoses of diseases, including a visual diagnosis of Tobacco rattle virus from South Carolina and sooty mold on peonies from Alaska, and physiological disorders, such as normal fall dieback from Canada, were made via email and using a Facebook page which as of the writing of this report has over 275 followers. Lastly, educational materials were created as a result of this project, namely, a workshop presented to the Alaska Peony Grower's Association and two presentations at the same conference. A journal article with the findings of this project is in review and a disease diagnosis bulletin is in preparation and pending submission. The information gained from this project will be valuable for growers and disease diagnosticians to make better diagnoses and subsequent disease management decisions.
Project objectives:
Objective 1. During the spring and summer of 2016 we will conduct surveys to obtain diseased foliage and solicit additional foliage to be collected by peony growers. Collections will focus on at least 30 growers in Alaska, Oregon, and Washington; however, they will extend to other Western states as identified during the course of the study.
Samples were collected from peonies from a total of 11 states: 1 farm in Connecticut, 2 farms in Maryland, 1 farm in New York, 1 farm in Pennsylvania, one farm in Virginia, 5 farms in Washington, 2 farms in Oregon, and 37 farms in Alaska and from landscape plantings in Indiana during the 2016 growing season. Surveys were also conducted on the plant material at the WSU Puyallup Research and Extension Center in Puyallup, WA. Samples processed from outside of the Western US were supported by another grant.
Objective 2. We will attempt to isolate causal organism of disease using traditional fungal and bacterial isolation methods. The isolated organisms will be maintained in-vitro in petri plates containing the appropriate growth media.
Isolations were performed to identify fungi or fungal-like organisms associated with disease symptoms. A total of 351 isolations were attempted by the graduate student and temporary employee from collected foliage using traditional fungal isolation techniques. Of these isolations, the majority were from Alaska (126), Washington (112), and Oregon (31). Figure 1 shows a comprehensive list of the states surveyed and the number of samples processed per state. With the exception of a root sample infected with Sclerotium rolfsii, from which sclerotia were taken from the surface of the infected root, a small piece of tissue was excised from the margin of a lesion and was surface sterilized in a 1% NaOCl solution and rinsed twice in sterile water. Surface sterilized tissue samples were then plated onto potato dextrose agar amended with streptomycin and chloramphenicol (PDA+s/c) at 10 mg/L each. Subsequent growth was transferred to a new PDA+s/c plate.
Attempts to isolate bacteria from samples that were unsuccessful at isolating a fungal pathogen were attempted for samples from Alaska, however, putative bacterial pathogens were not isolated from any samples. Subsequent growth of organisms was transferred to maintain pure cultures.
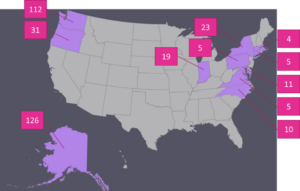
Figure 1. States from which samples of diseased peonies were collected during the 2016 growing season and the number of samples processed from each state.
Objective 3. We will use a combination of morphological and molecular methods (PCR and sequencing) to positively identify the suspected causal organisms of disease. If suspected pathogen is viral, samples will be submitted to a commercial virus screening laboratory for identification.
A combination of morphological and molecular techniques were used to identify fungal and fungal-like organism that were isolated from peony tissues. All fungal and fungal-like plant pathogens initially identified by morphology and PCR and sequencing of the internal transcribed spacer (ITS). DNA from isolates was extracted from isolates and used in PCR and sequencing of the ITS region of nuclear ribosomal DNA using primer pairs ITS6 and ITS4. Further identification by sequencing of the glyceraldehyde 3-phosphate dehydrogenase and the cytochrome c oxidase subunit 1 genes was used for the Colletotrichum and Sclerotinia and Phytophthora isolates, respectively. Sequences were compared with known sequences of fungal organisms published in the NCBI BLAST database. The sequences for all isolates used in pathogenicity trials were deposited into GenBank. Morphological traits used to identify fungi included colony and spore morphology and the production of resting structures. All fungi were identified to at least the genus level, with some identified to the species level.
Fungi and fungal-like organisms from 10 genera, plus at least one unidentified powdery mildew species were isolated and identified by morphology from the peony samples collected (Table 1). These pathogens were associated with a range of symptoms including leaf spots, stem spots or cankers, shoot blight, and root rot (Figure 2). The samples included five genera never before reported on peonies in the United States: a Botryosphaeria species, multiple Colletotrichum species, Mycocentrospora acerina, a Phoma species, and Pilidium concavum. Additional novel host-pathogen-state combinations were identified as described in Table 1. The most common disease identified on peonies, as determined by the number of states from which the disease was identified, was Botrytis gray mold caused by Botrytis spp. The second most common disease was measles, caused by Graphiopsis chlorocephala.

Samples of virus diseases were not collected during this study, thus, were not submitted to virus indexing. We were able to punitively diagnose Tobacco rattle virus on peonies in South Carolina and Alaska based on visual symptoms from photos provided by growers using the Facebook page developed as an outreach portion of this grant.
Table 1. Fungal and fungal-like diseases associated with peony by state in surveys conducted from 2013-2016. Diseases in bold represent first reports in the state in which they were found.
|
State |
Disease (causal organism)x |
|
Alaska |
Botrytis gray mold (multiple Botrytis spp.) Leaf spot (Sclerotinia sclerotiorum) Phoma (Phoma spp.)y Licorice spot [proposed, see discussion] (Mycocentrospora acerina)y |
|
Connecticut |
Anthracnose (Colletotrichum spp.)y |
|
Indiana |
Botrytis gray mold (unidentified Botrytis spp.)z Measles (Graphiopsis chlorocephala) Powdery mildew (unidentified spp.)z |
|
Maryland |
Anthracnose (Colletotrichum spp.)y Botrytis gray mold (unidentified Botrytis spp.)z Measles (Graphiopsis chlorocephala) White stem rot (Sclerotinia sclerotiorum) |
|
Michigan |
Leaf spot (unidentified Alternaria spp.)z |
|
Missouri |
Leaf spot (unidentified Alternaria spp.)z |
|
New York |
Botrytis gray mold (unidentified Botrytis spp.)z Leaf spot (Botryosphaeria spp.)y Leaf spot (unidentified Alternaria spp.) Powdery mildew (unidentified spp.)z |
|
North Carolina |
Botrytis gray mold (unidentified Botrytis spp.) Tan-brown leaf spot (Pilidium concavum)y White stem rot (Sclerotinia sclerotiorum) |
|
Oregon |
Anthracnose (Colletotrichum spp.)y Botrytis gray mold (multiple Botrytis spp.) Tan-brown leaf spot (Pilidium concavum)y Leaf spot (unidentified Alternaria spp.) Root rot (Phytophthora cactorum) |
|
Pennsylvania |
Leaf spot (unidentified Alternaria spp.)z Measles (Graphiopsis chlorocephala) Powdery mildew (unidentified spp.)z |
|
Virginia |
Tan-brown leaf spot (Pilidium concavum)y Measles (Graphiopsis chlorocephala) Powdery mildew (unidentified spp.)z |
|
Washington |
Anthracnose (Colletotrichum spp.)y Botrytis gray mold (multiple Botrytis spp.) Tan-brown leaf spot (Pilidium concavum)y Leaf spot (unidentified Alternaria spp.) Measles (Graphiopsis chlorocephala) Powdery mildew (unidentified spp.)z Licorice spot [proposed, see discussion] (Mycocentrospora acerina)y Southern blight (Sclerotium rolfsii) |
xDisease name is followed by causal organism in parentheses.
yIndicates pathogen represents first report in the United States.
zIndicates pathogenicity trials not conducted in this study.
Objective 4. We will perform Koch’s postulates on live plants and detached peony tissues to determine pathogenicity of the organisms.
For those fungal species that represented first reports on peony either in the United States or in the state in which the original sample was collected, pathogenicity trials were conducted using a representative isolate for each state-pathogen combination (Table 2). For all trials, isolates were grown up on PDA at 20C in the dark for 3-40 days, depending on species, with the exception of the Phytophthora cactorum which was grown up on V8 agar. Peony leaves, stems, or roots (depending on the site of original pathogen recovery) were removed from potted peony plants and surface sterilized in a 1% NaOCl solution for 3-5 minutes and then rinsed twice in sterile water. Plugs 5mm in diameter were cut from the actively growing region of the fungal colony and placed mycelium side down on the host tissue. For trials that took place on foliage, plugs were inoculated onto the abaxial side of the leaf. Both wounded and unwounded tissues were inoculated for most fungal species trials except for those that remained unwounded according to the results of preliminary pathogenicity trials and those conducted on stems and roots which were all wounded by pins used to hold plugs securely onto host tissue. For all trials, an uncolonized plug was used as a control. Inoculated and mock inoculated tissues were placed into a plastic bin containing wire racks suspended over moistened paper towels. A lid was placed on the bin and the bin was placed in a plastic bag to maintain high humidity conditions. Inoculated tissues were incubated at 20C under dark conditions. After sufficient observable symptom development (3-40 days depending on fungal species), plant material was removed from the incubator and the fungus was re-recovered from the tissue as described in the original fungal isolation protocol above. The identity of the re-isolated pathogen was confirmed by morphological analysis. A total of five replicates were performed per treatment. Most trials were conducted using the peony cultivar ‘Sarah Bernhardt.’ Trials for Graphiopsis chlorocephala were conducted on peony cultivar ‘Kansas’ due to the prior observation that this cultivar appeared highly susceptible to this pathogen, and those for a Colletotrichum spp. isolate from Connecticut took place on both ‘Sarah Bernhardt’ and ‘Kansas’ due to observations of varying susceptibility during pathogenicity trials.
Pathogenicity trials confirmed the ability of all organisms identified as first reports (Tables 1 and 2) to cause disease on peony consisting of symptoms like those observed on the original sample (Figure 2). A list of the isolates used for pathogenicity trials including GenBank accession numbers can be found in Table 2. In most cases, 5 of 5 inoculated tissues (wounded and unwounded) displayed symptoms, with successful re-isolation from the majority of lesions (data not shown). Lesions only developed on 4 of the 5 replicates for both wounded an unwounded tissues for the Colletotrichum spp. isolate from Oregon (HAM02) and the Botryosphaeria spp. isolate from New York (NY12). Lesions were produced on only 4 of 5 unwounded tissues for the Alternaria isolate from Washington (FR01) (pathogenicity tests were not conducted on wounded tissues). The Colletotrichum spp. isolate from Connecticut (CT03) appeared to have differences in virulence depending on peony cultivar. On ‘Kansas,’ CT03 was able to cause lesions characteristic of anthracnose infections on 4 of 5 wounded tissues and 2 of 5 unwounded tissues after 20 days, whereas symptoms only developed on 2 of 5 wounded and 1 of 5 unwounded tissues after 40 days. Lesions developed on tissue regardless of wounding for all isolates and wounding did not appear to influence the frequency of infection.
Table 2. Fungi isolated from diseased peonies and used in pathogenicity trials by state. All isolates were sequenced and sequences deposited in GenBank.
|
State |
Species |
Isolate Code |
Plant tissuez |
GenBank Accession Number(s) |
|
AK |
Sclerotinia sclerotiorum |
CC04 |
Foliage |
MF776031 |
|
Mycocentrospora acerina |
PE06 |
Stem |
MF776041 |
|
|
Phoma spp. |
RR06 |
Stem |
MF776043 |
|
|
CT |
Colletotrichum spp. |
CT03 |
Foliage |
MF776032; MF780699 |
|
IN |
Graphiopsis chlorocephala |
IN03 |
Foliage |
MF776048 |
|
MD |
Graphiopsis chlorocephala |
SF01c |
Flower bud |
MF776044 |
|
Colletotrichum spp. |
SF02b |
Stem |
MF776045; MF780702 |
|
|
NC |
Pilidium concavum |
NC10 |
Foliage |
MF776037 |
|
NY |
Alternaria spp. |
NY12b |
Foliage |
MF776039 |
|
Botryosphaeria spp. |
NY12 |
Foliage |
MF776038 |
|
|
OR |
Alternaria spp. |
ADE02 |
Foliage |
MF776028 |
|
Colletotrichum spp. |
HAM02 |
Foliage |
MF776036; MF780700 |
|
|
Pilidium concavum |
OP83 |
Foliage |
MF776040 |
|
|
Graphiopsis chlorocephala |
OR78b |
Stem |
MF776050 |
|
|
Phytophthora cactorum |
OP94 |
Tuberous root |
MF776049; MF780698 |
|
|
VA |
Pilidium concavum |
VA04 |
Foliage |
MF776047 |
|
Graphiopsis chlorocephala |
VA02 |
Foliage |
MF776046 |
|
|
WA |
Alternaria spp. |
FR01 |
Foliage |
MF776035 |
|
Sclerotium rolfsii |
DG92 |
Tuberous root |
MF776034 |
|
|
Pilidium concavum |
AR92 |
Foliage |
MF776030 |
|
|
Mycocentrospora acerina |
AR61 |
Stem |
MF776029 |
|
|
Colletotrichum spp. |
PUY45c |
Flower bud |
MF776042; MF780701 |
|
|
Graphiopsis chlorocephala |
WBC12 |
Stem |
MF776051 |
zPlant tissue from which the pathogen was originally isolated.
Objective 5. We will prepare and disseminate information regarding the peony pathogens. Such educational material will include an extension bulletin published through Washington State University and/or the University of Alaska Fairbanks, a workshop conducted in Alaska, and first reports published in peer-reviewed journals.
As a result of this project, two extension bulletins, one peer-reviewed paper, and one workshop have been prepared. One extension bulletin on Tobacco rattle virus has been published in collaboration with the University of Alaska Fairbanks, as described in a future section, and one on disease diagnosis and management is pending submission. A peer-reviewed journal article has been submitted to the journal Plant Disease and is in review. A workshop on general disease management and disease management in peonies was conducted in 2017 at the Alaska Peony Grower's Association Conference and is described in a future section. In addition, we conducted over 50 field visits to do field collections and meet with growers to discuss surveying for disease and disease diagnosis and management.