Final report for GNC21-332
Project Information
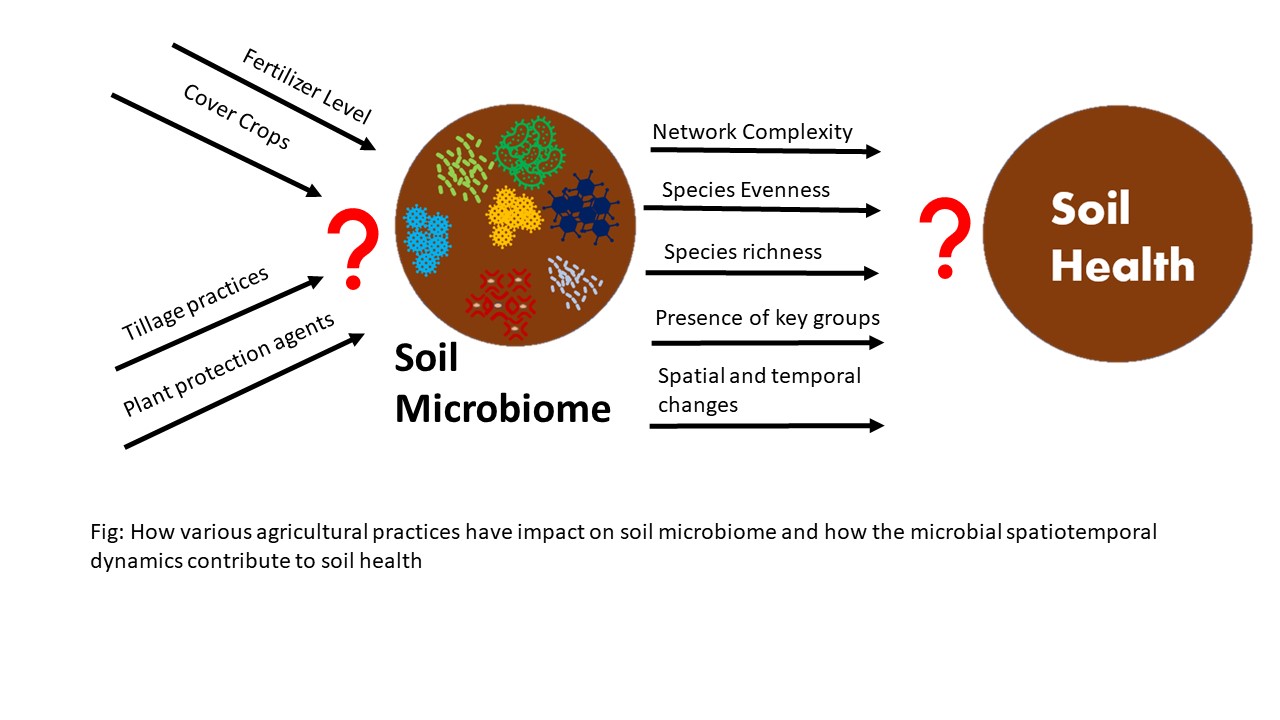
Growing global population demands for the increment on the food production on the same rate the population is increasing. Various agricultural production practices these days to attain these demands are threat for the food security of the future generation and for the balance of agroecosystems. The physical, chemical, and biological wellness of soil is important for the ecosystem balance and food production. Soil microbes play a pivotal role on the agroecosystem by facilitating number of processes incurring. Despite this, the role of microbial communities on the improvement of soil health has not been studied to optimize these benefits for the food production. This gap exists because of the inadequate understanding about the microbial communities and how they can be manipulated for sustainable agricultural practices that the future is seeking for. The clear definition and understanding of soil health and the role of soil microbiome on maintaining better soil health is still not vivid among the producers. The proposed research aims to investigate how microbial communities contribute to soil health and how agricultural practices have an impact on microbial spatiotemporal dynamics. In this project, I will use the North Dakota Agriculture Microbiome project as the baseline and extend it to conduct field surveys and manipulative experiments to understand the contribution of microbiomes to soil health and agricultural practices affect that contribution. I will perform the latest molecular sequencing approaches to analyze the assembly, recruitment and functioning of microbial communities under various agricultural practices. I will also test findings in a long-term field trial to further assess the consistency of field data. By combining the crop yield and management information from farmlands and the long-term field trial, I will create statistical models illustrating the contribution of microbial parameters to soil health under different scenarios. The knowledge on the importance of soil microbes and soil health improvement is growing among the producers of North Dakota. I will facilitate the deepening of this understanding by reaching the producers through existing extension activities. I will prepare the extension materials focusing the impact of the intensive agricultural practices have on the microbial communities and sustainable practices that can be adapted to promote these communities. The project aims to disseminate information about the microbial communities, their impact on plant and soil health, and the effects that agricultural practices have on these communities.
The project aims at exploring the impact various agricultural practices have on the spatiotemporal dynamics of microbes which includes their functioning, assembly, recruitment, and relationship within the microbiome. As an outcome, the project aims at an increased understanding of the producers of North Dakota about the soil health and roles of microbial communities in soil health. Further, the project aims to engage the producers in surveys so that they can learn how different agricultural practices shape microbiomes with subsequent impact on soil health. In addition, the project aims at preparing and publishing various extension materials which would help the producers gather knowledge on microbial communities at their agricultural lands. With this knowledge the project intends to help farmers adapt farming practices which would promote the soil health and adapt the sustainable approaches of farming as per the recommendations made. The project aims to increase awareness among the producers about the sustainable farming approaches that harness the soil microbes and improve overall physical, chemical, and biological qualities of soil. The project also aims to publish the scientific publications and impart them to the extension specialists of North Dakota, who acknowledge the producers about the production sustainability and soil wellness. Thus, the project aims to promote sustainable farming and climate-resilient agroecosystems in North Dakota.
Research
Bulk Soil and Plant Sampling
In August 2021 and August 2022, soil and plant samples were gathered from agricultural lands spanning North Dakota and Minnesota. In the first year, samples were taken from a total of 61 farms, including 31 conventional and 30 organic ones. Similarly, in the second year, samples were collected from 57 of the same farms, with 30 being conventional and 27 organics. USDA (United States Department of Agriculture) defines organic farming as a method that utilizes cultural, biological, and mechanical techniques to maintain on-farm resources, preserve the environment, and protect the variety of living organisms enhancing the ecosystem. (https://www.ams.usda.gov/sites/default/files/media/Organic%20Practices%20Factsheet.pdf). Conventional farms apply resources like fertilizer, water for irrigation, amendments, and pesticides evenly, without considering the natural variations in soil and crop conditions between and within fields. This leads to an uneven distribution of resources, with some areas receiving too much and others too little (Corwin & Scudiero, 2019). To prevent potential edge effects, the sampling was conducted at 25 meters from the edges. To ensure that the samples represented the entire farm, the sampling locations were chosen randomly. 5 soil cores (15 cm deep) were collected from the area around randomly chosen 5 plants resulting in a total of 25 soil cores from each of the 61 farms using a 1” diameter soil step probe. In 2021, 5 fully matured corn plants were uprooted from each farm using a shovel, and the same process was repeated in 2022 for the crop that was present at that time. The soil cores from each farm were collected in one-gallon Ziploc© bags which were then stored on ice packs in coolers during transportation to the laboratory. Once in the lab, plant particles and macro-organisms were removed from the soil and the soil cores were broken up and homogenized by sieving with a 2 mm sieve, and then divided into two 15 ml conical tubes. One of the tubes and the remaining soil in Ziplock bags were stored at -20°C for soil analysis, and the other was stored at -80°C for long-term storage for molecular analysis. In the field, the roots of the plants were clipped off at the root-shoot junction using sterilized secateurs, and 5 roots were collected and stored in a Ziplock bag and placed in coolers with ice packs during transportation to the laboratory. Once in the lab, the roots were washed with tap water to remove the soil. They were then washed in 95% EtOH for a minute and disinfected with a 10% bleach solution for a minute. Roots were again washed with 95% EtOH for 30 seconds and then rinsed with sterile water for 2 minutes. Excess water was removed using paper towels out of the processed roots. The roots were then cut into approximately 1cm pieces using scalpels, with about 40% of the cut roots from each plot stored in a 15 ml conical tube at -80°C for molecular analysis. Thinner roots were selected for mycorrhizal slide preparation and were placed in 15 ml conical tubes with 70% ethyl alcohol for longer storage. To measure the weight of fresh plants, they were wrapped and taped individually before being weighed. The collected plants were placed in brown paper bags and stored on ice packs for transporting to the lab. The plants were then dried in an oven at 40°C for approximately for 5 days to measure dry biomass.
Measurement of Soil and Plant Properties
100gm of bulk soil was measured from the soil samples held in Ziplock bags and placed in sealable bags provided by soil and plant biomass analyzing facility “AgVise Laboratories” located at Grand Forks, North Dakota for the analysis of the physiochemical properties of the soil. 50 gm of dried plant biomass was grinded and sent off to " AgVise Laboratories" for plant physiochemical properties analysis.
Mycorrhizal Colonization Assessment
Processed roots were further washed using one ml of a one M solution of (NaPO3)6, to remove any excess soil that remained. Approximately 15 root pieces were removed from each 15 ml conical tubes and kept into a 1.5 ml microcentrifuge tube. Labeling was done accordingly. (NaPO3)6 was added to each 1.5 ml microcentrifuge tubes, and the closed tubes were shaken for two hours. After two hours, the roots were removed and washed in deionized water. A 40% w/v KOH solution was added to the roots, and the mixture were incubated at 80°C for 30-40 minutes. After that, the roots were again rinsed with deionized water. Then, a 5% ink-vinegar solution was added to the roots and kept for 30-40 minutes at 80°C for staining. For the ink-vinegar stock solution, 50 ml ink (black from Parker Quink) was mixed with 950 ml of household vinegar (5% acetic acid). After the staining, the roots were rinsed with tap water and a drop of vinegar (Vierheilig et al., 1998). The stained roots were aligned parallel to the long axis of a microscopy slide. On each slide, there were five rows of roots- each having two to three pieces. Roots were fixed into place using 50% glycerol and a cover slip. The edges of the cover slips were securely sealed with coats of clear nail polish. The arbuscular mycorrhizal colonization was observed under a compound microscope at a magnification of 40X. The spot on the root surface where the center of the horizontal and vertical crosshairs entered through the side of the root was taken as the point of intersection. All intersections between the roots and the vertical crosshair were taken into consideration. Several categories were made to count the intersections: ‘arbuscules’, ‘vesicles’, ‘arbuscules and vesicles’, ‘only hyphae’, and ‘none’ i.e., there is no fungal material present in the root. To assess each intersection, the plane of focus was moved completely through the root. If the vertical crosshair cut one or more arbuscules or vesicles or hyphae, the appropriate category was incremented by one. For the arbuscular and vesicular colonization, the sum of the corresponding category was divided by the total number of intersections studied. The hyphal colonization was estimated as the ratio of non-negative intersections. One hundred intersections were calculated per sample to get the representative numbers (McGonigle et al., 1990b).
Microbiome analysis
The soil and root samples were frozen at -20°C until DNA extraction. To extract the DNA, 200 mg of soil was used for each bulk soil sample. For each root sample, 350 mg of root pieces were used. We utilized the Qiagen DNeasy Power Soil Pro kit and Qiagen DNeasy Plant Pro kit (Qiagen Inc., Valencia, CA) to extract the DNA, following the manufacturer’s instructions. We assessed the quality, quantity, and yield of the DNA using a NanoDrop ND-1000 spectrophotometer. After quantification, each DNA sample was normalized using nuclease-free water to a final volume of 50 µl at a concentration of 5 ng/µl. Standardized DNA samples have been sent to the University of Minnesota Genomics Center for amplicon sequencing for further microbiome analysis.
The results we have received so far are preliminary. However, we still need to conduct an extensive analysis using the microbiome data we are looking forward to.
Educational & Outreach Activities
Participation summary:
Last year, I participated in a NCB ASM (North Central Branch American Society of Microbiological Sciences) meeting where I presented preliminary results and progress of the project. As the project is ongoing for four years, we anticipate discovering fascinating results in the future.
2023/24 Update:
The project has engaged farmers and agricultural professionals through various educational activities and resources. We have regularly contacted 31 farmers before sampling their field and collected information on their farm management activities. During these calls, some afrmers discuss the importance of microbial communities in sustainable farming practices. We are currently preparing 1 journal article to share our research outcomes with the wider scientific community. I gave a talk during my thesis dissertation to share insights with graduate students, undergraduate researchers, professors, and research scientists who were interested in agriculture and microbiome. 40+ people were in that talk. My thesis can be found here which is entirely based on this project : https://www.proquest.com/openview/e5ff44364a5fb5d27a1ee2d19b627784/1?pq-origsite=gscholar&cbl=18750&diss=y . A field day is planned to engage local farmers directly, allowing them to learn about our findings. Moving forward, we have plans for additional presentations at conferences and an ongoing commitment to sharing knowledge through various outreach methods, including an upcoming tour of the Carrington Research Extension Center (CREC) to showcase our research initiatives. Our efforts aim to enhance awareness and understanding of sustainable agricultural practices among farmers and agricultural professionals.Final Defense sb this is the presentation I used in my final dissertation.
Project Outcomes
I believe this project has made significant impact toward enhancing agricultural sustainability by focusing on the role of microbial communities in soil health. Economically, our findings demonstrate that utilizing organic fertilization can improve soil quality and, consequently, crop yields, potentially reducing costs associated with chemical inputs and pest control. By identifying specific microbial families that act as biomarkers for organic practices, farmers can better manage their soil ecosystems, leading to increased productivity and profitability. Environmentally, the project highlights the benefits of organic fertilization in promoting biodiversity within soil communities. Our research shows that organic practices lead to higher bacterial diversity in the rhizosphere, contributing to healthier soil ecosystems that can better withstand abiotic stresses and reduce the need for synthetic fertilizers. This approach fosters a more balanced nutrient cycle, which is crucial for long-term soil fertility and sustainability. Socially, the project actively engages farmers through outreach activities such as field days, creating a platform for knowledge exchange and collaboration. By empowering farmers with insights into sustainable practices and the importance of microbial health, we foster a community that is better equipped to make informed decisions regarding their farming methods. Overall, this project promotes immediate benefits for farmers and also lays the groundwork for a more sustainable agricultural future in North Dakota and beyond.
Project Outcomes:
1. For bacteria, we noticed higher diversity in the rhizosphere, lower diversity in the roots, with intermediate levels of diversity in the bulk soil, for both conventional and organic treatments. While soil compartments influenced the bacterial community diversity very significantly (p<0.001), fertilization type had no influence on the diversity (p>0.05).
2. The fungal community diversity, akin to bacteria, were significantly different between compartments (p<0.001), while not significantly influenced by fertilization type (p>0.05). However, contrary to bacterial communities, we see lower levels of fungal diversity in the rhizosphere, and higher levels in the bulk soil, with roots showing intermediate levels.
3. Both bacterial and fungal communities are strongly structured by fertilization as well compartments (PERMANOVA p < 0.01).
4. Bacterial families such as Micrococcaceae and Xiphinematobacteraceae in the bulk soil, Rhizobiaceae and Oxalobacteraceae in the rhizosphere, Rhizobiaceae and Pseudonocardiaceae in the roots, were notable biomarker species for organic fertilization (at p<0.001).
5. Fungal families such as Microascaceae and Ascobolaceae in the bulk soil, Ceratobasidiaceae and Glomerellaceae in the rhizosphere, Hyaloscyphaceae and Microascaceae in the roots, were notable biomarker species for organic fertilization (at p<0.001).
Overall, our project has allowed us to observe shifts in microbial communities that happen under organic and conventional fertilization, in spatial resolution spanning bulk soil to rhizosphere and roots, over geographically separated fields, to arrive at robust microbial ecological conclusions.
During this project, my advisor and I significantly enhanced our knowledge and awareness of sustainable agriculture, particularly regarding its impact on microbial communities. We gained insights into how different fertilization methods influence microbial diversity and learned to identify key microbial families as biomarkers for organic practices. Our hands-on experience improved our skills in experimental design and data analysis, while our attitude became more proactive as we recognized the importance of advocating for soil health practices. Engaging directly with farmers deepened our understanding of their needs and concerns, motivating us to tailor our educational resources effectively and contribute positively to promoting sustainable agricultural practices within our community.