Final report for GNE19-215
Project Information
Pathogenic L. monocytogenes may inhabit dairy processing environments, increasing the risk for cross-contamination of foods. Using biocontrol microorganisms that inhibit or outcompete L. monocytogenes to complement sanitation of dairy processing facilities may enhance the control of L. monocytogenes. However, it remains unknown whether the resident microbiota of dairy processing facilities affects the antilisterial activity of biocontrol strains. Here, two lactic acid bacteria (LAB) strains (Lactococcs lactis subsp. lactis and Enterococcus durans) were tested for their biocontrol potential in the context of microbiomes collected from three ice cream processing facilities (A, B, and C). Antilisterial ability was assessed by co-culturing LABs with 8-L. monocytogenes strains in the presence of microbiota for 3 days at 15°C, followed by quantification of the most probable number of attached L. monocytogenes. The use of LABs did not significantly reduce the concentration of L. monocytogenes, compared to a positive control containing microbiota and L.monocytogenes without LAB. Specifically, L. monocytogenes concentration increased by 0.38±0.77 log10 MPN/sample in treatments containing microbiota from facility A, while it decreased by 0.99±1.13 and 2.54±0.84 log10 MPN/sample in treatments with microbiota from facilities B and C, respectively. The attachment of LAB to an abiotic surface was assessed by co-culturing LABs in with the microbiomes at 15°C for 3 days, followed by characterization of attached microbiota composition using amplicon sequencing. All samples containing microbiomes from facilities A and B had high relative abundance of Pseudomonas, while samples with facility C microbiome had high relative. abundance of Enterococcus. While we did not observe a significant reduction of L. monocytogenes within our model system, our work shows that the microbiota composition of ice cream processing facilities affected the antilisterial ability of LABs. Further studies are needed to evaluate whether certain taxa effectively inhibit or reduce the antilisterial properties of lactic acid bacteria, and the mechanisms behind microbiota interactions. In addition, we developed two educational videos on the topics of "Listeria in the dairy industry" and "Biofilms in the dairy industry" which will be publised along extension articles, and be made available for dairy processors and used during outreach workshops.
Objective 1: Characterization of small-scale dairy processing facility microbiomes.
Objective 2: Measure the ability of protective cultures to effectively colonize small-scale dairy environment microbiome biofilms.
Objective 3: Test the efficacy of protective cultures against Listeria monocytogenes (Lm) within an in vitro grown dairy environment microbiome biofilm.
Objective 4: Development of educational materials for dissemination of results among stakeholders.
Pennsylvania produces over 15% of the total milk supply in the U.S. and 99% of Pennsylvania dairy farms are family owned. Many of them are small farms that process the milk they produce to add value to their product. However, previous extension research from Penn State indicates that small-scale dairy processors face food safety challenges that can compromise their businesses. One of the main microbiological food safety hazards associated with the dairy supply chain is Listeria monocytogenes (Lm). The Center for Disease Control and Prevention estimates that out of 1,591 cases of human listeriosis occurring every year in the United States, 260 result in death. Lm has been associated with dairy foods, including soft cheeses, ice cream and raw milk. The presence of Lm in ready-to-eat food may result in recalls that generate an economic loss to producers, as well as distrust of their brand’s image. It has been estimated that the annual cost of listeriosis in the United States 2.6 billion dollars.
While Lm is effectively inactivated during pasteurization, it may be re-introduced to food products when present in the food processing environment. It is therefore critical for food processors to effectively control Lm presence in the environment to prevent contamination of products and maintain consumer trust. However, it is particularly challenging to eradicate Lm from food processing environments, since it can survive and grow in cold and humid environments, such as dairy processing facilities. While Lm is susceptible to common cleaning and sanitizing practices applied by the dairy industry, it can grow in biofilms with bacterial communities present in the processing environments. Biofilms are attached aggregates of bacteria that secrete slime-like extracellular polymeric substances that bind them together and protect them from environmental stressors such as cleaners and sanitizers. Lm can benefit from the interactions with certain environmental microbiomes, such as Pseudomonas, since Pseudomonas is a very good biofilm former that can provide protection to Lm. Nevertheless, these interactions with environmental microbiome can also be leveraged in food processors’ advantage, if protective cultures that can outcompete or inhibit the growth of biofilm formers and Lm are added to the environment. It is known that certain microbial species, including lactic acid bacteria, can suppress the growth and adhesion of Lm. I therefore propose to test two biocontrol strains that had previously been shown to be effective in inhibition of Lm in poultry processing facilities. If successful, implementation of such protective cultures in the food processing environment may provide enhanced benefits compared to sanitizers alone, since biocontrol strains have the added advantage of not promoting antimicrobial tolerance and resistance development in Lm and will likely be effective also against antimicrobial-resistant Lm strains. If the results show merit in our in vitro study, these interventions may be tested in the future in actual facility environments. his approach is considered sustainable, because it is based on natural interactions among beneficial microorganisms that can be leveraged in small-scale dairy processors’ advantage. Specifically, application of protective biocontrol cultures is less likely to facilitate the development of Lm resistance, which often occurs as a result of chemical treatments.
While I am not seeking to replace traditional sanitation methods, I believe biological control can be use in addition to physical and chemical interventions to enhance their efficacy. This project will be the first to test the efficacy of protective cultures in conjunction with dairy facility microbiome. If successful, I will continue to investigate the application of biocontrol agents within an actual dairy processing facility. In addition to research, I intend to develop educational materials focused on environmental control of Lm. These will communicate the importance of cleaning and sanitation practices to reduce Lm incidence in food processing environments, as well as explain how microbiome biofilms can contribute to persistence of Lm in food processing facilities. It also will be important to inform the end users in how biological control interventions should be without compromising the facility hygienic status. Overall, I believe this project is an important first step towards providing small-scale processors with new strategies for Lm control, resulting in enhanced sustainability of their business through improved food safety.
Cooperators
- (Educator and Researcher)
- (Educator and Researcher)
Research
Objective 1: Characterization of small-scale dairy processing facility microbiome.
1.1 Sample collection. Three small-scale ice cream processing facilities were enrolled in this study. These facilities are small to medium-size operations, that produce a variety of dairy products, including milk, cheese, and ice cream. Six samples were collected from each facility using 3M hydrated sponges with neutralizing buffer by a combination of ten horizontal and 10 vertical swabbing moves form an area of 40 cm by 40 cm or an equivalent area in sites that are not flat. Sampled areas included non-food contact surfaces such as drains and floors among others, within the ice cream processing room in each facility. Samples were stored on ice in a cooler during the transportation to the lab. During sampling, the temperature of each dairy processing facility was measured to adjust the incubation temperature of our in vitro experiments (Obj. 2 and 3) to represent dairy processing facility conditions. Ninety milliliters of Brain Heart Infusion (BHI) broth were added to each of the six samples collected per facility. Samples were homogenized using a laboratory stomacher for 14 minutes at 230rpm, to assure that all soil was transferred to the media, and homogenates of all samples were combined to obtain a microbiome sample representative of each facility. Twenty percent glycerol v/v was added to homogenates to allow for preservation at -80 °C.
1.2 DNA extraction and 16S rDNA extraction for microbiome sequencing. Fifty milliliters of the homogenate prepared in 1.1. were transferred to a sterile 50 ml conical tube and centrifuged at 11,000 g and 4 ºC for 20 minutes. After centrifugation, the supernatants were discarded and pellets containing microbiomes were stored at -80 °C until DNA extraction. DNA was extracted from approximately 0.25 g sample using Omega E.Z.N.A Soil DNA kit. A sterile sponge (representative of a production lot) homogenized in BHI was also processed, as a negative control, to confirm absence of microbial DNA contaminants in the sterile sponge and BHI. Concentration of DNA after extraction was determined with Nanodrop One and using Qubit dsDNA High Sensitivity Assay Kit. PCR was used to amplify the16S rDNA V4 region of the bacteria present in each sample. Briefly, the V4 region of the 16S rRNA gene sequence was amplified using a forward primer 505F-v2 (TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG GTG YCA GCM GCC GCG GTA A) and a reverse primer 806R-v2 (GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGG ACT ACN VGG GTW TCT AAT). The PCR mastermix was composed of 12.5ul of KAPA 2x KAPA HIFI HotStart Ready Mix, 1 μl of 10 μM forward primer, 1μl of 10 μM reverse primer and 8.5 μl of nuclease-free watering 2 μl of the extracted DNA were added per reaction. A positive control (microbiome sample from an apple packing house that was obtained following the same sampling protocol) was included in each run, as well as a negative control with 2 μl of nuclease free water. PCR thermal cycling for amplification of the 16S rRNA gene V4 region was conducted as follows: initial denaturation at 98ºC for 2 min, 30 cycles of denaturation at 98ºC for 20 s, annealing at 60ºC for 20 s, extension at 72ºC for 25 s, and final extension at 72ºC for 5 min; final hold was at 4ºC. The results of the PCR reactions were visualized on a 2 % agarose gel electrophoresis run at 130V for 30 minutes to confirm successful amplification. Concentration of DNA amplicons will be determined by Qubit 3 using Qubit dsDNA High Sensitivity Assay Kit. Extracted DNA was sent to an external laboratory (Wright Labs or Novogene) for PCR amplification and sequencing of the 16S rDNA V4 region of the bacteria present in each sample.
1.3 Bioinformatics. Sequences were analyzed with DADA2 pipeline following the standard protocol for 16S rRNA V4 region amplicon sequence reads. Paired-end sequencing reads were merged into contigs and contigs shorter or longer than 254 bp were discarded. Chimera were detected and removed. Amplicon sequence variants (ASVs) were assigned taxonomy by aligned against the reference database Silva (version 132).
1.4 Statistical analyses of microbiome data. To avoid eliminating a subset of data through normalization or rarefaction, we used the microbiome compositional data analysis approach. All ASVs with “0” count value were assigned a small, non-zero value using the Count Zero Multiplicative Method with the zCompositions package (version 1.3.4) prior to applying the center-log ratio (CLR) transformation. To visualize the data, Principal Component Analysis (PCA) was used on CLR-transformed data. To explore the taxonomic compositions of samples, relative abundances of taxa were calculated using the Aitchison Simplex method with the R package compositions (version 1.40-5). The composition of microbiota for each facility was visualized at the family taxonomic level using stacked bar plots. Bacterial ASVs with less than 2% relative abundance were merged into the category “Others” and bar plots were generated with the package ggplot2 (version 3.3.2).
Objective 2: Measure the ability of protective cultures to effectively colonize small-scale dairy environment microbiome biofilms.
2.1 Putative protective cultures. Lactic acid bacteria (LAB) strains Lactococcus lactis ssp. lactis C-1-92 (LL) and Enterococcus durans 152 (ED) were obtained from the American Type Culture Collection (ATCC).
2.2 Colonization of protective cultures in reconstituted dairy facility microbiome. To determine whether the two LAB strains can attach to a model surface (i.e., polypropylene) in the presence of microbiota collected from ice cream processing facilities, we added them to aliquots of composite microbiota suspensions. The two LAB cultures were added to microbiota each individually, as well as together. For each facility, the microbiota suspension prepared in 1.1 was aliquoted into 15-ml polypropylene conical tubes in duplicates. Tubes were inoculated with each LAB strain at a final concentration of 5*107 CFU/ml to reach a total volume of 2 ml. Negative controls were not inoculated with the LAB strains to measure the effect of LAB strains on the composition of attached microbiota. Inoculated tubes were incubated at 15°C for 3 days to allow for attachment cells to the surface. The temperature of incubation was defined as the mean temperature measured in dairy processing facilities during sampling at the time of sample collection. After incubation, the medium was removed from tubes, and the attached biomass was washed twice with 0.1% peptone water to remove planktonic and loosely attached cells. The attached microbiota was resuspended in 2 ml of 0.1% peptone water by adding 2 g of 3 mm sterile glass beads and vortexing for 30 seconds. Ten-fold serial dilutions of resuspended microbiota were prepared in 0.1 % peptone water and spread plated in triplicate onto (BHI) agar to quantify the total amount of culturable bacteria and De Man, Rogosa, and Sharpe (MRS) agar to quantify LAB. The inoculated plates were incubated at 35°C for 3 days. Each experiment had two technical replicates per treatment and was conducted in two independent replicates. One milliliter of the resuspended attached microbiota suspension was used to characterize the microbial composition of attached biomass. DNA from the resuspended biomass was extracted using the DNeasy Power Biofilm kit (Qiagen, Germantown, MD). The extracted DNA was used for PCR-amplification of the 16S rRNA V4 region, library preparation, sequencing and analysis as described in 1.2 – 1.4.
Objective 3: Measure the ability of protective cultures to inhibit L. monocytogenes in the presence of small-scale dairy environment microbiome biofilms.
3.1 Listeria monocytogenes isolates. Eight Listeria monocytogenes strains of different linages that were previously isolated from dairy processing environments, were obtained from Dr. Martin Weidmann at Cornell University.
3.2 Inhibition of L. monocytogenes by protective cultures in reconstituted dairy facility microbiome. To determine whether the two LAB strains can effectively inhibit L. monocytogenes when in the presence of the microbiota collected from ice cream processing facilities, we added them to aliquots of the microbiome suspensions, individually and combined. The microbiota suspension collected from each facility was prepared as outlined in 1.1 and was aliquoted into 15-ml conical polypropylene tubes in duplicate. Tubes were inoculated with each biocontrol strain at a final concentration of 5*107 CFU/ml, and with of an 8-strain cocktail of L. monocytogenes at a final concentration of 3*106 CFU/ml. Negative controls were not inoculated with the LAB strains or L. monocytogenes to measure the effect of the strains on the microbial composition of attached biomass. Positive controls were inoculated with L. monocytogenes without the addition of LAB strains. The tubes were incubated at 15°C for 3 days to allow for the attachment of cells to the surface. After incubation, the medium was removed, and the attached biomass was washed twice in 0.1% peptone water to remove planktonic and loosely attached cells. Attached cells were resuspended in 2 ml of 0.1% peptone water by adding 2 g of 3 mm sterile glass beads and vortexing for 30 seconds. Ten-fold serial dilutions of resuspended attached biomass were prepared in 0.1% peptone water and were spread plated in triplicate onto BHI agar. Inoculated plates were incubated at 35°C for 3 days. To quantify the concentration of L. monocytogenes in the attached biomass, a modified FDA BAM MPN protocol was used. For each treatment and the initial environmental microbiota suspension, 7-decimal dilutions were made in 0.1% peptone water. Then, 100 μl of each dilution were transferred to three microcentrifuge tubes containing 900 μl of Buffered Listeria Enrichment broth (BLEB), followed by incubation at 30°C for 4 hours. After incubation, 4 μl of Selective Listeria Enrichment Supplement (90 mg acriflavine, 450 mg cycloheximide and 360 mg sodium nalidixic acid in 40 ml of sterile DI water) was added to each tube followed by incubation at 30°C for 44 additional hours. After incubation, an aliquot of each tube was streaked onto Agar Listeria Ottavani & Agosti (ALOA) and incubated at 37°C for 48 hours. The presence of blue colonies with a halo indicated the presence of L. monocytogenes in the enrichment. The number of positive L. monocytogenes MPN tubes was recorded, and the final concentration was calculated using the MPN calculator available from the FDA BAM appendix 2: Most Probable Number from Serial Dilutions. The experiment was conducted in two independent biological replicates. One milliliter of the attached biomass suspension was used to characterize its bacterial composition. DNA from the attached biomass was extracted using the DNeasy Power Biofilm kit (Qiagen, Germantown, MD). Extracted DNA was used to PCR-amplification of the 16S rRNA V4 region, library preparation, sequencing and analysis as described in 1.2 – 1.4.
Objective 4: Development of educational and outreach videos.
Educational videos on the topics of "Listeria in the dairy industry" and "Biofilms in the dairy industry" were developed and will be made available to stakeholders via the Penn State Extension website. In addition, two extension articles were written to include with each video to provide additional information to dairy processors and additional resources. Both videos and extension articles were translated to Spanish.
Objective 1: Characterization of small-scale dairy processing facility microbiome.
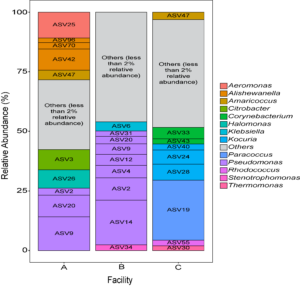
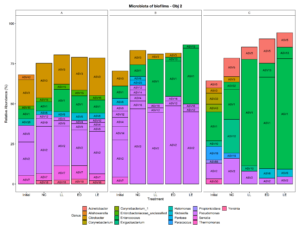
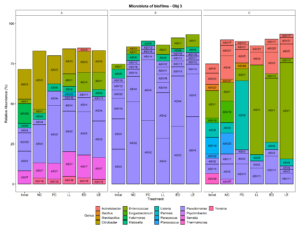
Three small scale ice cream processing facilities were enrolled to participate in this study. The facilities are coded as "A", "B", and "C" throughout the report to preserve confidentiality. Six samples were collected in each facility from non-food contact surfaces including floors and drains within the ice cream processing environment of each facility and combined into one composite sample for microbiome characterization. To characterize and compare the composition of the microbiota between ice cream processing facilities, we analyzed the sequencing data using the compositional data analysis approach. Center-log ratio transformation was performed to the ASV table. To visualize the taxonomic composition of the microbiota of the ice cream processing environments, we examined the relative abundance of ASVs that were present over 2 % relative abundance (Fig. 1). Aeromonas (10.9 % - ASV25), Alishewanella (13.7 % - ASV 96, ASV70, ASV42), Amaricoccus (4.1 % - ASV47), Citrobacter (8.7 % - ASV3), Halomonas (7.7 % - ASV26), and Pseudomonas (26.5 % - ASV2, ASV20, ASV9) were the most prevalent genera in Facility A; Klebsiella (4.0 % - ASV6), Pseudomonas (48.7 % - ASV31, ASV20,ASV9, ASV12, ASV4, ASV2, ASV14) and Stenotrophomonas (2.4 % - ASV34) were the most prevalent genera in Facility B; and Amaricoccus (3.2 % - ASV47), Corynebacterium (7.1 % - ASV33, ASV43), Kocuria (15.5 % - ASV40, ASV24, ASV28), Paracoccus (25.7 % - ASV19), Rhodococcus (2.4 % - ASV55) and Thermomonas (2.1 % - ASV30) in Facility C (Fig. 1).
Objective 2: Measure the ability of protective cultures to effectively colonize small-scale dairy environment microbiome biofilms.
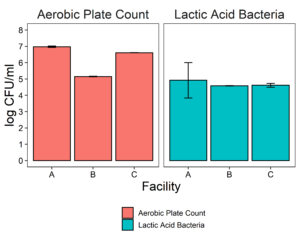
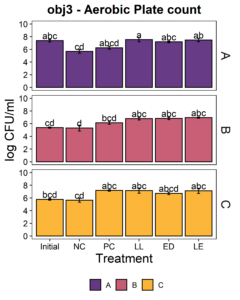
The putative protective cultures of LAB, Lactococcus lactis subsp. lactis (LL) C-1-92 and Enterococcus durans (ED) 152, that have been shown to successfully control Listeria monocytogenes in drains of a poultry processing facility, were obtained from ATCC. To measure whether these LAB cultures can attach to an abiotic surface in the presence of the microbiota of small-scale ice cream processing facilities, we co-cultured the microbiota with each strain independently and together. We first quantified the amount of culturable organisms present in the composite microbiome samples of each facility (Fig. 2) with an aerobic plate count (APC) method. Total aerobic bacteria counts represent all the bacteria that can grow aerobically at the temperature of incubation, 15°C, while LAB count represent the bacteria that can grow on MRS medium, a selective medium for LAB. Microbiome samples collected from facilities A and C were not significantly different in their APC (Fac. A = 6.97 log10CFU/ml; Fac. C = 6.61 log10CFU/ml). Microbiome samples collected from facility B had a significantly APC than the other two facilities in this study (Fac. B = 5.14 log10CFU/ml). However, microbiomes collected from all facilities were not significantly different in their LAB count (Fac. A = 4.92 log10CFU/ml; Fac. B = 4.58 log10CFU/ml; Fac. C=4.61 log10CFU/ml).
Microbiota composite samples were allowed to attach to the polypropylene surface in the presence of LAB cultures for 3 days at 15°C. A negative control was included to determine whether there was an increase in the total aerobic bacteria count and lactic acid bacteria count when LAB strains were added to the environmental microbiome. In experiments using samples from all three facilities, there was a significant increase (p<0.05) in the total aerobic bacteria count and lactic acid bacteria count in attached biomass in samples with added LAB strains (Fig. 3). However, there was no significant difference between treatments, indicating that LAB cultures can attach with environmental microbiota collected from ice cream processing facilities, either each LAB strain individually (LL or ED) or combined (LL and ED).
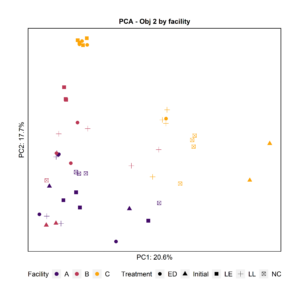
To further characterize the bacterial composition of the attached biomass and the initial microbiota used in the experiments, we extracted DNA and sequenced the 16S rRNA V4 gene region. Principal Component Analysis and Pairwise PERMANOVA showed that the attached biomass was significantly different among facilities (Fig. 4). When examining the effect of the treatment (LL, ED, or LE) on the attached microbiota for all facilities together, we observed that the initial microbiota added at day 0 was significantly different from the treatments LL and ED. Further, the negative control’s attached biomass was significantly different from samples with added LE and ED, and the LL treatment was significantly different from LE.
To visualize the microbiota composition, we plotted the mean relative abundance of all ASVs that were present in any treatment in a relative abundance above 2 % (Fig. 5). The most prevalent ASVs in Facility A were identified as strains of Pseudomonas and Enterobacteriaceae (Klebsiella, Citrobacter, and Serratia), regardless of the treatment. The most prevalent ASVs in Facility B were identified as strains of Pseudomonas, regardless of the treatment. The most prevalent ASVs in Facility C were identified as strains of Enterococcus, regardless of the treatment. ASV1 and ASV 13 (Enterococcus) likely represent the two LAB cultures added to the treatments. These results suggests that the bacteria that are present in the environmental microbiome of processing plants may have an effect the attachment of putative protective cultures added to the environment. Pseudomonas is a psychrotroph and a good biofilm former that may outcompete other bacteria when attaching to a surface, if present in high concentrations. Interestingly, in Facility C, which had an initial lower relative abundance of Pseudomonas ASVs, the LAB cultures were able to effectively attach to the surface and dominate the attached microbiota composition.
Objective 3: Test the efficacy of protective cultures against Listeria monocytogenes (Lm) within an in vitro grown dairy environment microbiome biofilm.
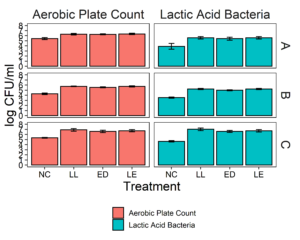
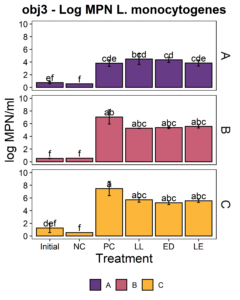
We aimed to determine whether the LAB strains L. lactis C-1-92 (LL) and E. durans 152 (ED) could inhibit L. monocytogenes in the presence of the environmental microbiota of ice cream processing facilities. For this purpose, microbiome composite samples were allowed to attach to polypropylene surface in the presence of LAB and an 8-strain cocktail of L. monocytogenes strains, for 3 days at 15°C. A positive control containing the microbiome and the L. monocytogenes cocktail was included to determine the reduction of L. monocytogenes due to the addition of the LAB strains. A negative control was included to determine whether there was an increase in the total aerobic bacteria count or L. monocytogenes concentration in test samples with added ED, LL, or both LL and ED (LE). To quantify the inhibition of L. monocytogenes in the attached microbiota, L. monocytogenes concentration using the MPN method. Further, we quantified the total culturable organisms in the attached biomass. For all facilities, there was a significant increase (p<0.05) in the total aerobic bacteria count in attached biomass in samples to which LAB strains were added (Fig. 6). However, there was no significant difference between LL, ED, and LE treatments. Compared to the positive control, there was no significant reduction of L. monocytogenes due to the addition of LL, ED, or LE in the presence of environmental microbiota collected from any of the three facilities (p>0.05).
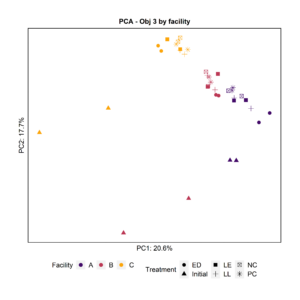
To further characterize the bacterial composition of the attached microbiota and the initial microbiota used in the experiments, we extracted DNA and sequenced the 16S rRNA V4 gene region. Principal Component Analysis and pairwise PERMANOVA indicated that the attached biomass was significantly different by facility (Fig. 8). The initial planktonic microbiome used at day 0 was significantly different from the attached microbiota after 3 days of incubation in all the treatments including the negative and positive controls.
To visualize the microbiota composition, the mean relative abundance of all ASVs that were present in any treatment at a relative abundance above 2% is plotted in Fig. 9. Similar to the results obtained for the prior experiment (Objective 2), the most prevalent ASVs were strains of Pseudomonas and Enterobacteriaceae (Klebsiella, Citrobacter, and Serratia) in Facility A, strains of Pseudomonas in Facility B, and strains of Enterococcus in Facility C, regardless of the treatment. ASV1 and ASV 13 (Enterococcus) likely represent the two LAB strains added to the treatments.
Project results were summarized in a manuscript entitled “Context matters: environmental microbiota of ice cream processing facilities affects the inhibitory performance of two lactic acid bacteria against Listeria monocytogenes”, which is under review in a scientific journal mSphere, and was additionally submitted to the preprint repository bioRxiv (https://www.biorxiv.org/content/10.1101/2022.10.04.510917v1)
Objective 4: Development of educational materials for dissemination of results among stakeholders.
We developed two scripts and animations for videos on the topics of Listeria contamination in dairy processing environments and the roles of biofilms/microbiomes in the dairy industry. Two extension articles were written to provide additional information and resources along with the videos. Videos and extension articles were sent to Penn State extension for publishing on their website (https://extension.psu.edu/), and we expect them to be uploaded by December 2022. In addition, we leveraged resources available through Penn State Extension, and we translated the videos and extension articles to Spanish, to increase our reach to dairy processors and industry personnel whose native language is not English.
The two lactic acid bacteria proposed as biological control agents in food processing environments need to compete for space and nutrients with the resident microbiota present in food processing facilities, which may affect their antilisterial activity. This study showed that the composition of the microbiota collected from ice cream processing facilities and the presence of certain microbial taxa affect the attachment and inhibitory effect of lactic acid bacteria strains against L. monocytogenes. Further studies are needed to evaluate whether certain taxa effectively inhibit or reduce the antilisterial properties of lactic acid bacteria, and the mechanisms behind microbiota interactions.
Education & Outreach Activities and Participation Summary
Participation Summary:
We developed two videos on the topics of "Listeria in the dairy industry" and "Biofilsm in the dairy industry". These materials will be posted in the Penn State Extension website along two extension articles that provide additional technical details to the information in the videos. Furthermore, the videos will be used during workshops and short courses focused on dairy foods safety. We were able to leverage resources from other funding sources to translate both videos and extension articles into Spanish, to increase our audience.
Project Outcomes
The use of biological controls during cleaning and sanitizing of food processing environments represents a sustanable approach to reduce the incidence of foodborne pathogens that may contaminate food. The outcomes of this project include experimental data showing potential effects of environmental microbiota in the effectiveness of two lactic acid bacteria that may be applied in dairy processing facilities to reduce the incidence of L. monocytogenes. Our results show that the composition and relative abundances of certain organisms present in the environments (such as Pseudomonas or Enterobacteriaceae), may affect the effectiveness of biological controls applied. Nonetheless, further work needs to be carried out to verify our results in the field, and to evaluate the mechanisms of action.
Over the course of this project I have acquired skills in molecular biology and methods to sample and characterize environmental microbiomes, including DNA extraction methods, sequencing and bioinformatics. I have also taken courses in Agriculture Social Change applied to international development. The concepts learned in these course can be applied in this project.
During my doctoral degree, I plan to continue to investigate on the role of environmental microbiomes regarding food safety of food processing facilities. I am looking forward to applying novel genomic techniques to enhance food safety in dairy processing plants as well as other food processing environments. Furthermore, I am interested in learning and applying advanced microbiological methods, including the use of molecular biology and environmental microbiology, to solve technical issues related tofood processing, quality and safety. I am particularly interested in generating tailored-solutions the development of sustainable food systems, and to mitigate food waste. I aim to become a scientific official for a development organization, such as the Food and Agricultural Organization, so that I may assist with developing sustainable food systems.
Information Products
- Biofilms in the Dairy Industry (Article/Newsletter/Blog)
- Listeria in the Dairy Industry (Article/Newsletter/Blog)
- Listeria in the Dairy Industry (Video)
- Biofilms in the Dairy Industry (Video)